The apop_data structure represents a data set. It joins together a gsl_vector, a gsl_matrix, an apop_name, and a table of strings. It tries to be lightweight, so you can use it everywhere you would use a gsl_matrix or a gsl_vector.
Here is a diagram showing a sample data set with all of the elements in place. Together, they represent a data set where each row is an observation, which includes both numeric and text values, and where each row/column may be named.
| Rowname | Vector | Matrix | Text | Weights | ||||||||||||||||||||||||||||||||||
|
|
|
|
|
In a regression, the vector would be the dependent variable, and the other columns (after factor-izing the text) the independent variables. Or think of the apop_data set as a partitioned matrix, where the vector is column -1, and the first column of the matrix is column zero. Here is some sample code to print the vector and matrix, starting at column -1 (but you can use apop_data_print to do this).
Most functions assume that each row represents one observation, so the data vector, data matrix, and text have the same row count: data->vector->size==data->matrix->size1 and data->vector->size==*data->textsize. This means that the apop_name structure doesn't have separate vector_names, row_names, or text_row_names elements: the rownames are assumed to apply for all.
See below for notes on managing the text element and the row/column names.
The apop_data set includes a more pointer, which will typically be NULL, but may point to another apop_data set. This is intended for a main data set and a second or third page with auxiliary information, such as estimated parameters on the front page and their covariance matrix on page two, or predicted data on the front page and a set of prediction intervals on page two.
The more pointer is not intended for a linked list for millions of data points. In such situations, you can often improve efficiency by restructuring your data to use a single table (perhaps via apop_data_pack and apop_data_unpack).
Most functions, such as apop_data_copy and apop_data_free, will handle all the pages of information. For example, an optimization search over multi-page parameter sets would search the space given by all pages.
Pages may also be appended as output or auxiliary information, such as covariances, and an MLE would not search over these elements. Any page with a name in XML-ish brackets, such as <Covariance>, is considered information about the data, not data itself, and therefore ignored by search routines, missing data routines, et cetera. This is achieved by a rule in apop_data_pack and apop_data_unpack.
Here is a toy example that establishes a baseline data set, adds a page, modifies it, and then later retrieves it.
There are a great many functions to collate, copy, merge, sort, prune, and otherwise manipulate the apop_data structure and its components.
Apophenia builds upon the GSL, but it would be inappropriate to redundantly replicate the GSL's documentation here. Meanwhile, here are prototypes for a few common functions. The GSL's naming scheme is very consistent, so a simple reminder of the function name may be sufficient to indicate how they are used.
gsl_matrix_swap_rows (gsl_matrix * m, size_t i, size_t j) gsl_matrix_swap_columns (gsl_matrix * m, size_t i, size_t j) gsl_matrix_swap_rowcol (gsl_matrix * m, size_t i, size_t j) gsl_matrix_transpose_memcpy (gsl_matrix * dest, const gsl_matrix * src) gsl_matrix_transpose (gsl_matrix * m) : square matrices only gsl_matrix_set_all (gsl_matrix * m, double x) gsl_matrix_set_zero (gsl_matrix * m) gsl_matrix_set_identity (gsl_matrix * m) gsl_matrix_memcpy (gsl_matrix * dest, const gsl_matrix * src) void gsl_vector_set_all (gsl_vector * v, double x) void gsl_vector_set_zero (gsl_vector * v) int gsl_vector_set_basis (gsl_vector * v, size_t i): set all elements to zero, but set item 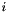 to one.
to one. gsl_vector_reverse (gsl_vector * v): reverse the order of your vector's elements gsl_vector_ptr and gsl_matrix_ptr. To increment an element in a vector use, e.g., *gsl_vector_ptr(v, 7) += 3; or (*gsl_vector_ptr(v, 7))++. gsl_vector_memcpy (gsl_vector * dest, const gsl_vector * src)The apop_text_to_data() function takes in the name of a text file with a grid of data in (comma|tab|pipe|whatever)-delimited format and reads it to a matrix. If there are names in the text file, they are copied in to the data set. See Input text file formatting for the full range and details of what can be read in.
If you have any columns of text, then you will need to read in via the database: use apop_text_to_db() to convert your text file to a database table, do any database-appropriate cleaning of the input data, then use apop_query_to_data() or apop_query_to_mixed_data() to pull the data to an apop_data set.
You may not need to use these functions often, given that apop_query_to_data, apop_text_to_data, and many transformation functions will auto-allocate apop_data sets for you.
The apop_data_alloc function allocates a vector, a matrix, or both. After this call, the structure will have blank names, NULL text element, and NULL weights. See Name handling for discussion of filling the names. Use apop_text_alloc to allocate the text grid. The weights are a simple gsl_vector, so allocate a 100-unit weights vector via allocated_data_set->weights = gsl_vector_alloc(100).
Examples of use can be found throughout the documentation; for example, see A quick overview.
See also:
gsl_matrix * gsl_matrix_alloc (size_t n1, size_t n2) gsl_matrix * gsl_matrix_calloc (size_t n1, size_t n2) void gsl_matrix_free (gsl_matrix * m) gsl_vector * gsl_vector_alloc (size_t n) gsl_vector * gsl_vector_calloc (size_t n) void gsl_vector_free (gsl_vector * v)There are several macros for the common task of viewing a single row or column of a apop_data set.
Because these macros can be used as arguments to a function, these macros have abbreviated names to save line space.
A second set of macros have a slightly different syntax, taking the name of the object to be declared as the last argument. These can not be used as expressions such as function arguments.
The view is an automatic variable, not a pointer, and therefore disappears at the end of the scope in which it is declared. If you want to retain the data after the function exits, copy it to another vector:
Curly braces always delimit scope, not just at the end of a function. When program evaluation exits a given block, all variables in that block are erased. Here is some sample code that won't work:
For this if/then statement, there are two sets of local variables generated: one for the if block, and one for the else block. By the last line, neither exists. You can get around the problem here by making sure to not put the macro declaring new variables in a block. E.g.:
First, some examples:
.row and a .rowname, I go with the name; similarly for .col and .colname. apop_data_get(dataset, 1) gets item (1, 0) from the matrix element of dataset. But as a do-what-I-mean exception, if there is no matrix element but there is a vector, then this form will get vector element 1. Relying on this DWIM exception is useful iff you can guarantee that a data set will have only a vector or a matrix but not both. Otherwise, be explicit and use apop_data_get(dataset, 1, -1).The apop_data_ptr function follows the lead of gsl_vector_ptr and gsl_matrix_ptr, and like those functions, returns a pointer to the appropriate double. For example, to increment the (3,7)th element of an apop_data set:
apop_data_(get|set|ptr). See also:
double gsl_matrix_get (const gsl_matrix * m, size_t i, size_t j) double gsl_vector_get (const gsl_vector * v, size_t i) void gsl_matrix_set (gsl_matrix * m, size_t i, size_t j, double x) void gsl_vector_set (gsl_vector * v, size_t i, double x) double * gsl_matrix_ptr (gsl_matrix * m, size_t i, size_t j) double * gsl_vector_ptr (gsl_vector * v, size_t i) const double * gsl_matrix_const_ptr (const gsl_matrix * m, size_t i, size_t j) const double * gsl_vector_const_ptr (const gsl_vector * v, size_t i) gsl_matrix_get_row (gsl_vector * v, const gsl_matrix * m, size_t i) gsl_matrix_get_col (gsl_vector * v, const gsl_matrix * m, size_t j) gsl_matrix_set_row (gsl_matrix * m, size_t i, const gsl_vector * v) gsl_matrix_set_col (gsl_matrix * m, size_t j, const gsl_vector * v)These functions allow you to send each element of a vector or matrix to a function, either producing a new matrix (map) or transforming the original (apply). The ..._sum functions return the sum of the mapped output.
There are two types, which were developed at different times. The apop_map and apop_map_sum functions use variadic function inputs to cover a lot of different types of process depending on the inputs. Other functions with types in their names, like apop_matrix_map and apop_vector_apply, may be easier to use in some cases. They use the same routines internally, so use whichever type is convenient.
You can do many things quickly with these functions.
Get the sum of squares of a vector's elements:
Create an index vector [  ].
].
Given your log likelihood function, which acts on a apop_data set with only one row, and a data set where each row of the matrix is an observation, find the total log likelihood via:
How many missing elements are there in your data matrix?
Get the mean of the not-NaN elements of a data set:
The following program randomly generates a data set where each row is a list of numbers with a different mean. It then finds the 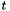 statistic for each row, and the confidence with which we reject the claim that the statistic is less than or equal to zero.
statistic for each row, and the confidence with which we reject the claim that the statistic is less than or equal to zero.
Notice how the older apop_vector_apply uses file-global variables to pass information into the functions, while the apop_map uses a pointer to send parameters to the functions.
One more toy example demonstrating the use of apop_map and apop_map_sum :
![$[0, 1]$](form_104.png) , et cetera
, et cetera See also:
int gsl_matrix_add (gsl_matrix * a, const gsl_matrix * b) int gsl_matrix_sub (gsl_matrix * a, const gsl_matrix * b) int gsl_matrix_mul_elements (gsl_matrix * a, const gsl_matrix * b) int gsl_matrix_div_elements (gsl_matrix * a, const gsl_matrix * b) int gsl_matrix_scale (gsl_matrix * a, const double x) int gsl_matrix_add_constant (gsl_matrix * a, const double x) gsl_vector_add (gsl_vector * a, const gsl_vector * b) gsl_vector_sub (gsl_vector * a, const gsl_vector * b) gsl_vector_mul (gsl_vector * a, const gsl_vector * b) gsl_vector_div (gsl_vector * a, const gsl_vector * b) gsl_vector_scale (gsl_vector * a, const double x) gsl_vector_add_constant (gsl_vector * a, const double x) matrix, matrix
matrix, matrix  vector, or vector
vector, or vector  matrix
matrix See the GSL documentation for myriad further options.
See also:
double gsl_matrix_max (const gsl_matrix * m) double gsl_matrix_min (const gsl_matrix * m) void gsl_matrix_minmax (const gsl_matrix * m, double * min_out, double * max_out) void gsl_matrix_max_index (const gsl_matrix * m, size_t * imax, size_t * jmax) void gsl_matrix_min_index (const gsl_matrix * m, size_t * imin, size_t * jmin) void gsl_matrix_minmax_index (const gsl_matrix * m, size_t * imin, size_t * jmin, size_t * imax, size_t * jmax) gsl_vector_max (const gsl_vector * v) gsl_vector_min (const gsl_vector * v) gsl_vector_minmax (const gsl_vector * v, double * min_out, double * max_out) gsl_vector_max_index (const gsl_vector * v) gsl_vector_min_index (const gsl_vector * v) gsl_vector_minmax_index (const gsl_vector * v, size_t * imin, size_t * imax)For most of these, you can add a weights vector for weighted mean/var/cov/..., such as apop_vector_mean(d->vector, .weights=d->weights)
There are no functions provided to convert from apop_data to the constituent elements, because you don't need a function.
If you need an individual element, you can use its pointer to retrieve it:
In the other direction, you can use compound literals to wrap an apop_data struct around a loose vector or matrix:
double* 
gsl_vector double*  apop_data
apop_data  apop_data
apop_data  database table
database table If you generate your data set via apop_text_to_data or from the database via apop_query_to_data (or apop_query_to_text or apop_query_to_mixed_data) then column names appear as expected. Set apop_opts.db_name_column to the name of a column in your query result to use that column name for row names.
Sample uses, given apop_data set d:
The apop_data set includes a grid of strings, named text, for holding text data.
Text should be encoded in UTF-8. ASCII is a subset of UTF-8, so that's OK too.
There are a few simple forms for handling the text element of an apop_data set.
tdata is tdata->textsize[0]; the number of columns is tdata->textsize[1]. tdata->text[row][col]. x[0] can always be written as *x, which may save some typing. The number of rows is *tdata->textsize. If you have a single column of text data (i.e., all data is in column zero), then item i is *tdata->text[i]. If you know you have exactly one cell of text, then its value is **tdata->text. "", which you can check via Here is a sample program that uses these forms, plus a few text-handling functions.
dataset = apop_query_to_text("select onecolumn from data"); then you have a sequence of strings, d->text[0][0], d->text[1][0], .... After apop_data dt = apop_data_transpose(dataset), you will have a single list of strings, dt->text[0], which is often useful as input to list-of-strings handling functions.Factor is jargon for a numbered category. Number-crunching programs prefer integers over text, so we need a function to produce a one-to-one mapping from text categories into numeric factors.
A dummy is a variable that is either one or zero, depending on membership in a given group. Some methods (typically when the variable is an input or independent variable in a regression) prefer dummies; some methods (typically for outcome or dependent variables) prefer factors. The functions that generate factors and dummies will add an informational page to your apop_data set with a name like <categories for your_column> listing the conversion from the artificial numeric factor to the original data. Use apop_data_get_factor_names to get a pointer to that page.
You can use the factor table to translate from numeric categories back to text (though you probably have the original text column in your data anyway).
Having the factor list in an auxiliary table makes it easy to ensure that multiple apop_data sets use the same single categorization scheme. Generate factors in the first set, then copy the factor list to the second, then run apop_data_to_factors on the second:
See the documentation for apop_logit for a sample linear model using a factor dependent variable and dummy independent variable.