Data Structures | |
| struct | apop_arms_settings |
| struct | apop_cdf_settings |
| struct | apop_composition_settings |
| struct | apop_coordinate_transform_settings |
| struct | apop_cross_settings |
| struct | apop_data |
| struct | apop_dconstrain_settings |
| struct | apop_kernel_density_settings |
| struct | apop_lm_settings |
| struct | apop_loess_settings |
| struct | apop_mcmc_proposal_s |
| struct | apop_mcmc_settings |
| struct | apop_mixture_settings |
| struct | apop_mle_settings |
| struct | apop_model |
| struct | apop_name |
| struct | apop_opts_type |
| struct | apop_parts_wanted_settings |
| struct | apop_pm_settings |
| struct | apop_pmf_settings |
| struct | apop_settings_type |
Macros | |
| #define | apop_ANOVA |
| #define | apop_ANOVA |
| #define | apop_ANOVA |
| #define | apop_ANOVA |
| #define | Apop_c(d, col) |
| #define | Apop_c(d, col) |
| #define | Apop_c(d, col) |
| #define | Apop_c(d, col) |
| #define | Apop_col_t(d, colname, outd) |
| #define | Apop_col_t(d, colname, outd) |
| #define | Apop_col_t(d, colname, outd) |
| #define | Apop_col_t(d, colname, outd) |
| #define | Apop_col_tv(m, col, v) |
| #define | Apop_col_tv(m, col, v) |
| #define | Apop_col_tv(m, col, v) |
| #define | Apop_col_tv(m, col, v) |
| #define | Apop_cs(d, colnum, len) |
| #define | Apop_cs(d, colnum, len) |
| #define | Apop_cs(d, colnum, len) |
| #define | Apop_cs(d, colnum, len) |
| #define | Apop_cv(data_to_view, col) |
| #define | Apop_cv(data_to_view, col) |
| #define | Apop_cv(data_to_view, col) |
| #define | Apop_cv(data_to_view, col) |
| #define | apop_data_add_names(dataset, type, ...) |
| #define | apop_data_add_names(dataset, type, ...) |
| #define | apop_data_add_names(dataset, type, ...) |
| #define | apop_data_add_names(dataset, type, ...) |
| #define | apop_data_falloc(sizes, ...) |
| #define | apop_data_falloc(sizes, ...) |
| #define | apop_data_falloc(sizes, ...) |
| #define | apop_data_falloc(sizes, ...) |
| #define | apop_data_fill(adfin, ...) |
| #define | apop_data_fill(adfin, ...) |
| #define | apop_data_fill(adfin, ...) |
| #define | apop_data_fill(adfin, ...) |
| #define | apop_data_free(freeme) |
| #define | apop_data_free(freeme) |
| #define | apop_data_free(freeme) |
| #define | apop_data_free(freeme) |
| #define | apop_data_prune_columns(in, ...) |
| #define | apop_data_prune_columns(in, ...) |
| #define | apop_data_prune_columns(in, ...) |
| #define | apop_data_prune_columns(in, ...) |
| #define | apop_errorlevel |
| #define | apop_errorlevel |
| #define | apop_errorlevel |
| #define | apop_errorlevel |
| #define | apop_estimate_r_squared(in) |
| #define | apop_estimate_r_squared(in) |
| #define | apop_estimate_r_squared(in) |
| #define | apop_estimate_r_squared(in) |
| #define | apop_F_distribution |
| #define | apop_F_distribution |
| #define | apop_F_distribution |
| #define | apop_F_distribution |
| #define | apop_F_test |
| #define | apop_F_test |
| #define | apop_F_test |
| #define | apop_F_test |
| #define | apop_gaussian |
| #define | apop_gaussian |
| #define | apop_gaussian |
| #define | apop_gaussian |
| #define | apop_IV |
| #define | apop_IV |
| #define | apop_IV |
| #define | apop_IV |
| #define | Apop_mcv(matrix_to_view, col) |
| #define | Apop_mcv(matrix_to_view, col) |
| #define | Apop_mcv(matrix_to_view, col) |
| #define | Apop_mcv(matrix_to_view, col) |
| #define | apop_mean |
| #define | apop_mean |
| #define | apop_mean |
| #define | apop_mean |
| #define | apop_model_coordinate_transform(...) |
| #define | apop_model_coordinate_transform(...) |
| #define | apop_model_coordinate_transform(...) |
| #define | apop_model_coordinate_transform(...) |
| #define | apop_model_copy_set(model, type, ...) |
| #define | apop_model_copy_set(model, type, ...) |
| #define | apop_model_copy_set(model, type, ...) |
| #define | apop_model_copy_set(model, type, ...) |
| #define | apop_model_cross(...) |
| #define | apop_model_cross(...) |
| #define | apop_model_cross(...) |
| #define | apop_model_cross(...) |
| #define | apop_model_dcompose(...) |
| #define | apop_model_dcompose(...) |
| #define | apop_model_dcompose(...) |
| #define | apop_model_dcompose(...) |
| #define | apop_model_dconstrain(...) |
| #define | apop_model_dconstrain(...) |
| #define | apop_model_dconstrain(...) |
| #define | apop_model_dconstrain(...) |
| #define | apop_model_mixture(...) |
| #define | apop_model_mixture(...) |
| #define | apop_model_mixture(...) |
| #define | apop_model_mixture(...) |
| #define | apop_model_set_parameters(in, ...) |
| #define | apop_model_set_parameters(in, ...) |
| #define | apop_model_set_parameters(in, ...) |
| #define | apop_model_set_parameters(in, ...) |
| #define | Apop_model_set_settings(model, ...) |
| #define | Apop_model_set_settings(model, ...) |
| #define | Apop_model_set_settings(model, ...) |
| #define | Apop_model_set_settings(model, ...) |
| #define | apop_model_set_settings |
| #define | apop_model_set_settings |
| #define | apop_model_set_settings |
| #define | apop_model_set_settings |
| #define | Apop_mrv(matrix_to_view, row) |
| #define | Apop_mrv(matrix_to_view, row) |
| #define | Apop_mrv(matrix_to_view, row) |
| #define | Apop_mrv(matrix_to_view, row) |
| #define | Apop_notify(verbosity, ...) |
| #define | Apop_notify(verbosity, ...) |
| #define | Apop_notify(verbosity, ...) |
| #define | Apop_notify(verbosity, ...) |
| #define | apop_OLS |
| #define | apop_OLS |
| #define | apop_OLS |
| #define | apop_OLS |
| #define | apop_PMF |
| #define | apop_PMF |
| #define | apop_PMF |
| #define | apop_PMF |
| #define | Apop_r(d, rownum) |
| #define | Apop_r(d, rownum) |
| #define | Apop_r(d, rownum) |
| #define | Apop_r(d, rownum) |
| #define | apop_rng_get_thread(thread_in) |
| #define | apop_rng_get_thread(thread_in) |
| #define | apop_rng_get_thread(thread_in) |
| #define | apop_rng_get_thread(thread_in) |
| #define | Apop_row_t(d, rowname, outd) |
| #define | Apop_row_t(d, rowname, outd) |
| #define | Apop_row_t(d, rowname, outd) |
| #define | Apop_row_t(d, rowname, outd) |
| #define | Apop_row_tv(m, row, v) |
| #define | Apop_row_tv(m, row, v) |
| #define | Apop_row_tv(m, row, v) |
| #define | Apop_row_tv(m, row, v) |
| #define | Apop_rs(d, rownum, len) |
| #define | Apop_rs(d, rownum, len) |
| #define | Apop_rs(d, rownum, len) |
| #define | Apop_rs(d, rownum, len) |
| #define | Apop_rv(data_to_view, row) |
| #define | Apop_rv(data_to_view, row) |
| #define | Apop_rv(data_to_view, row) |
| #define | Apop_rv(data_to_view, row) |
| #define | Apop_settings_add_group(model, type, ...) |
| #define | Apop_settings_add_group(model, type, ...) |
| #define | Apop_settings_add_group(model, type, ...) |
| #define | Apop_settings_add_group(model, type, ...) |
| #define | Apop_settings_copy(name, ...) |
| #define | Apop_settings_copy(name, ...) |
| #define | Apop_settings_copy(name, ...) |
| #define | Apop_settings_copy(name, ...) |
| #define | Apop_settings_declarations(ysg) |
| #define | Apop_settings_declarations(ysg) |
| #define | Apop_settings_declarations(ysg) |
| #define | Apop_settings_declarations(ysg) |
| #define | Apop_settings_free(name, ...) |
| #define | Apop_settings_free(name, ...) |
| #define | Apop_settings_free(name, ...) |
| #define | Apop_settings_free(name, ...) |
| #define | Apop_settings_get(model, type, setting) |
| #define | Apop_settings_get(model, type, setting) |
| #define | Apop_settings_get(model, type, setting) |
| #define | Apop_settings_get(model, type, setting) |
| #define | Apop_settings_get_group(m, type) |
| #define | Apop_settings_get_group(m, type) |
| #define | Apop_settings_get_group(m, type) |
| #define | Apop_settings_get_group(m, type) |
| #define | Apop_settings_init(name, ...) |
| #define | Apop_settings_init(name, ...) |
| #define | Apop_settings_init(name, ...) |
| #define | Apop_settings_init(name, ...) |
| #define | Apop_settings_rm_group(m, type) |
| #define | Apop_settings_rm_group(m, type) |
| #define | Apop_settings_rm_group(m, type) |
| #define | Apop_settings_rm_group(m, type) |
| #define | Apop_settings_set(model, type, setting, data) |
| #define | Apop_settings_set(model, type, setting, data) |
| #define | Apop_settings_set(model, type, setting, data) |
| #define | Apop_settings_set(model, type, setting, data) |
| #define | Apop_stopif(test, onfail, level, ...) |
| #define | Apop_stopif(test, onfail, level, ...) |
| #define | Apop_stopif(test, onfail, level, ...) |
| #define | Apop_stopif(test, onfail, level, ...) |
| #define | Apop_subm(matrix_to_view, srow, scol, nrows, ncols) |
| #define | Apop_subm(matrix_to_view, srow, scol, nrows, ncols) |
| #define | Apop_subm(matrix_to_view, srow, scol, nrows, ncols) |
| #define | Apop_subm(matrix_to_view, srow, scol, nrows, ncols) |
| #define | apop_sum |
| #define | apop_sum |
| #define | apop_sum |
| #define | apop_sum |
| #define | apop_test_ANOVA_independence(d) |
| #define | apop_test_ANOVA_independence(d) |
| #define | apop_test_ANOVA_independence(d) |
| #define | apop_test_ANOVA_independence(d) |
| #define | apop_text_fill(dataset, ...) |
| #define | apop_text_fill(dataset, ...) |
| #define | apop_text_fill(dataset, ...) |
| #define | apop_text_fill(dataset, ...) |
| #define | apop_var |
| #define | apop_var |
| #define | apop_var |
| #define | apop_var |
| #define | apop_vector_fill(avfin, ...) |
| #define | apop_vector_fill(avfin, ...) |
| #define | apop_vector_fill(avfin, ...) |
| #define | apop_vector_fill(avfin, ...) |
Functions | |
| apop_data * | apop_anova (char *table, char *data, char *grouping1, char *grouping2) |
| int | apop_arms_draw (double *out, gsl_rng *r, apop_model *m) |
| gsl_vector * | apop_array_to_vector (double *in, int size) |
| apop_model * | apop_beta_from_mean_var (double m, double v) |
| apop_data * | apop_bootstrap_cov (apop_data *data, apop_model *model, gsl_rng *rng, int iterations, char keep_boots, char ignore_nans, apop_data **boot_store) |
| double | apop_cdf (apop_data *d, apop_model *m) |
| void | apop_crosstab_to_db (apop_data *in, char *tabname, char *row_col_name, char *col_col_name, char *data_col_name) |
| void | apop_data_add_named_elmt (apop_data *d, char *name, double val) |
| void | apop_data_add_names_base (apop_data *d, const char type, char const **names) |
| apop_data * | apop_data_add_page (apop_data *dataset, apop_data *newpage, const char *title) |
| apop_data * | apop_data_alloc (const size_t size1, const size_t size2, const int size3) |
| apop_data * | apop_data_calloc (const size_t size1, const size_t size2, const int size3) |
| apop_data * | apop_data_copy (const apop_data *in) |
| apop_data * | apop_data_correlation (const apop_data *in) |
| apop_data * | apop_data_covariance (const apop_data *in) |
| apop_data * | apop_data_fill_base (apop_data *in, double[]) |
| char | apop_data_free_base (apop_data *freeme) |
| double | apop_data_get (const apop_data *data, size_t row, int col, const char *rowname, const char *colname, const char *page) |
| apop_data * | apop_data_get_factor_names (apop_data *data, int col, char type) |
| apop_data * | apop_data_get_page (const apop_data *data, const char *title, const char match) |
| apop_data * | apop_data_listwise_delete (apop_data *d, char inplace) |
| void | apop_data_memcpy (apop_data *out, const apop_data *in) |
| gsl_vector * | apop_data_pack (const apop_data *in, gsl_vector *out, char more_pages, char use_info_pages) |
| apop_data * | apop_data_pmf_compress (apop_data *in) |
| void | apop_data_print (const apop_data *data, Output_declares) |
| void | apop_data_print (const apop_data *data, char const *output_name, FILE *output_pipe, char output_type, char output_append) |
| apop_data * | apop_data_prune_columns_base (apop_data *d, char **colnames) |
| double * | apop_data_ptr (apop_data *data, int row, int col, const char *rowname, const char *colname, const char *page) |
| apop_data * | apop_data_rank_compress (apop_data *in, int min_bins) |
| apop_data * | apop_data_rank_expand (apop_data *in) |
| void | apop_data_rm_columns (apop_data *d, int *drop) |
| apop_data * | apop_data_rm_page (apop_data *data, const char *title, const char free_p) |
| apop_data * | apop_data_rm_rows (apop_data *in, int *drop, int(*do_drop)(apop_data *, void *), void *drop_parameter) |
| int | apop_data_set (apop_data *data, size_t row, int col, const double val, const char *rowname, const char *colname, const char *page) |
| void | apop_data_show (const apop_data *data) |
| apop_data * | apop_data_sort (apop_data *data, apop_data *sort_order, char asc, char inplace, double *col_order) |
| apop_data ** | apop_data_split (apop_data *in, int splitpoint, char r_or_c) |
| apop_data * | apop_data_stack (apop_data *m1, apop_data *m2, char posn, char inplace) |
| apop_data * | apop_data_summarize (apop_data *data) |
| apop_data * | apop_data_to_bins (apop_data const *indata, apop_data const *binspec, int bin_count, char close_top_bin) |
| int | apop_data_to_db (const apop_data *set, const char *tabname, char) |
| apop_data * | apop_data_to_dummies (apop_data *d, int col, char type, int keep_first, char append, char remove) |
| apop_data * | apop_data_to_factors (apop_data *data, char intype, int incol, int outcol) |
| apop_data * | apop_data_transpose (apop_data *in, char transpose_text, char inplace) |
| void | apop_data_unpack (const gsl_vector *in, apop_data *d, char use_info_pages) |
| int | apop_db_close (char vacuum) |
| int | apop_db_open (char const *filename) |
| apop_data * | apop_db_to_crosstab (char const *tabname, char const *row, char const *col, char const *data, char is_aggregate) |
| double | apop_det_and_inv (const gsl_matrix *in, gsl_matrix **out, int calc_det, int calc_inv) |
| apop_data * | apop_dot (const apop_data *d1, const apop_data *d2, char form1, char form2) |
| int | apop_draw (double *out, gsl_rng *r, apop_model *m) |
| apop_model * | apop_estimate (apop_data *d, apop_model *m) |
| apop_data * | apop_estimate_coefficient_of_determination (apop_model *) |
| void | apop_estimate_parameter_tests (apop_model *est) |
| apop_model * | apop_estimate_restart (apop_model *e, apop_model *copy, char *starting_pt, double boundary) |
| apop_data * | apop_f_test (apop_model *est, apop_data *contrast) |
| long double | apop_generalized_harmonic (int N, double s) |
| apop_data * | apop_histograms_test_goodness_of_fit (apop_model *h0, apop_model *h1) |
| apop_data * | apop_jackknife_cov (apop_data *data, apop_model *model) |
| long double | apop_kl_divergence (apop_model *from, apop_model *to, int draw_ct, gsl_rng *rng) |
| long double | apop_linear_constraint (gsl_vector *beta, apop_data *constraint, double margin) |
| double | apop_log_likelihood (apop_data *d, apop_model *m) |
| apop_data * | apop_map (apop_data *in, apop_fn_d *fn_d, apop_fn_v *fn_v, apop_fn_r *fn_r, apop_fn_dp *fn_dp, apop_fn_vp *fn_vp, apop_fn_rp *fn_rp, apop_fn_dpi *fn_dpi, apop_fn_vpi *fn_vpi, apop_fn_rpi *fn_rpi, apop_fn_di *fn_di, apop_fn_vi *fn_vi, apop_fn_ri *fn_ri, void *param, int inplace, char part, int all_pages) |
| apop_data * | apop_map (apop_data *in, double(*fn_d)(double), double(*fn_v)(gsl_vector *), double(*fn_r)(apop_data *), double(*fn_dp)(double, void *), double(*fn_vp)(gsl_vector *, void *), double(*fn_rp)(apop_data *, void *), double(*fn_dpi)(double, void *, int), double(*fn_vpi)(gsl_vector *, void *, int), double(*fn_rpi)(apop_data *, void *, int), double(*fn_di)(double, int), double(*fn_vi)(gsl_vector *, int), double(*fn_ri)(apop_data *, int), void *param, int inplace, char part, int all_pages) |
| double | apop_map_sum (apop_data *in, apop_fn_d *fn_d, apop_fn_v *fn_v, apop_fn_r *fn_r, apop_fn_dp *fn_dp, apop_fn_vp *fn_vp, apop_fn_rp *fn_rp, apop_fn_dpi *fn_dpi, apop_fn_vpi *fn_vpi, apop_fn_rpi *fn_rpi, apop_fn_di *fn_di, apop_fn_vi *fn_vi, apop_fn_ri *fn_ri, void *param, char part, int all_pages) |
| double | apop_map_sum (apop_data *in, double(*fn_d)(double), double(*fn_v)(gsl_vector *), double(*fn_r)(apop_data *), double(*fn_dp)(double, void *), double(*fn_vp)(gsl_vector *, void *), double(*fn_rp)(apop_data *, void *), double(*fn_dpi)(double, void *, int), double(*fn_vpi)(gsl_vector *, void *, int), double(*fn_rpi)(apop_data *, void *, int), double(*fn_di)(double, int), double(*fn_vi)(gsl_vector *, int), double(*fn_ri)(apop_data *, int), void *param, char part, int all_pages) |
| void | apop_matrix_apply (gsl_matrix *m, void(*fn)(gsl_vector *)) |
| void | apop_matrix_apply_all (gsl_matrix *in, void(*fn)(double *)) |
| gsl_matrix * | apop_matrix_copy (const gsl_matrix *in) |
| double | apop_matrix_determinant (const gsl_matrix *in) |
| gsl_matrix * | apop_matrix_inverse (const gsl_matrix *in) |
| int | apop_matrix_is_positive_semidefinite (gsl_matrix *m, char semi) |
| gsl_vector * | apop_matrix_map (const gsl_matrix *m, double(*fn)(gsl_vector *)) |
| gsl_matrix * | apop_matrix_map_all (const gsl_matrix *in, double(*fn)(double)) |
| double | apop_matrix_map_all_sum (const gsl_matrix *in, double(*fn)(double)) |
| double | apop_matrix_map_sum (const gsl_matrix *in, double(*fn)(gsl_vector *)) |
| double | apop_matrix_mean (const gsl_matrix *data) |
| void | apop_matrix_mean_and_var (const gsl_matrix *data, double *mean, double *var) |
| apop_data * | apop_matrix_pca (gsl_matrix *data, int const dimensions_we_want) |
| void | apop_matrix_print (const gsl_matrix *data, Output_declares) |
| void | apop_matrix_print (const gsl_matrix *data, char const *output_name, FILE *output_pipe, char output_type, char output_append) |
| gsl_matrix * | apop_matrix_realloc (gsl_matrix *m, size_t newheight, size_t newwidth) |
| void | apop_matrix_show (const gsl_matrix *data) |
| gsl_matrix * | apop_matrix_stack (gsl_matrix *m1, gsl_matrix const *m2, char posn, char inplace) |
| long double | apop_matrix_sum (const gsl_matrix *m) |
| double | apop_matrix_to_positive_semidefinite (gsl_matrix *m) |
| void | apop_maximum_likelihood (apop_data *data, apop_model *dist) |
| apop_model * | apop_ml_impute (apop_data *d, apop_model *meanvar) |
| apop_model * | apop_model_clear (apop_data *data, apop_model *model) |
| apop_model * | apop_model_copy (apop_model *in) |
| apop_model * | apop_model_cross_base (apop_model *mlist[]) |
| apop_data * | apop_model_draws (apop_model *model, int count, apop_data *draws) |
| long double | apop_model_entropy (apop_model *in, int draws) |
| apop_model * | apop_model_fix_params (apop_model *model_in) |
| apop_model * | apop_model_fix_params_get_base (apop_model *model_in) |
| void | apop_model_free (apop_model *free_me) |
| apop_data * | apop_model_hessian (apop_data *data, apop_model *model, double delta) |
| apop_model * | apop_model_metropolis (apop_data *d, gsl_rng *rng, apop_model *m) |
| int | apop_model_metropolis_draw (double *out, gsl_rng *rng, apop_model *model) |
| apop_model * | apop_model_mixture_base (apop_model **inlist) |
| apop_data * | apop_model_numerical_covariance (apop_data *data, apop_model *model, double delta) |
| void | apop_model_print (apop_model *model, FILE *output_pipe) |
| apop_model * | apop_model_set_parameters_base (apop_model *in, double ap[]) |
| void | apop_model_show (apop_model *print_me) |
| apop_model * | apop_model_to_pmf (apop_model *model, apop_data *binspec, long int draws, int bin_count) |
| long double | apop_multivariate_gamma (double a, int p) |
| long double | apop_multivariate_lngamma (double a, int p) |
| int | apop_name_add (apop_name *n, char const *add_me, char type) |
| apop_name * | apop_name_alloc (void) |
| apop_name * | apop_name_copy (apop_name *in) |
| int | apop_name_find (const apop_name *n, const char *findme, const char type) |
| void | apop_name_free (apop_name *free_me) |
| void | apop_name_print (apop_name *n) |
| void | apop_name_stack (apop_name *n1, apop_name *nadd, char type1, char typeadd) |
| gsl_vector * | apop_numerical_gradient (apop_data *data, apop_model *model, double delta) |
| double | apop_p (apop_data *d, apop_model *m) |
| apop_data * | apop_paired_t_test (gsl_vector *a, gsl_vector *b) |
| apop_model * | apop_parameter_model (apop_data *d, apop_model *m) |
| apop_data * | apop_predict (apop_data *d, apop_model *m) |
| void | apop_prep (apop_data *d, apop_model *m) |
| int | apop_prep_output (char const *output_name, FILE **output_pipe, char *output_type, char *output_append) |
| int | apop_query (const char *q,...) |
| apop_data * | apop_query_to_data (const char *fmt,...) |
| double | apop_query_to_float (const char *fmt,...) |
| apop_data * | apop_query_to_mixed_data (const char *typelist, const char *fmt,...) |
| apop_data * | apop_query_to_text (const char *fmt,...) |
| gsl_vector * | apop_query_to_vector (const char *fmt,...) |
| apop_data * | apop_rake (char const *margin_table, char *const *var_list, int var_ct, char *const *contrasts, int contrast_ct, char const *structural_zeros, int max_iterations, double tolerance, char const *count_col, char const *init_table, char const *init_count_col, double nudge) |
| int | apop_regex (const char *string, const char *regex, apop_data **substrings, const char use_case) |
| gsl_rng * | apop_rng_alloc (int seed) |
| gsl_rng * | apop_rng_get_thread_base (int thread) |
| double | apop_rng_GHgB3 (gsl_rng *r, double *a) |
| void | apop_score (apop_data *d, gsl_vector *out, apop_model *m) |
| int | apop_system (const char *fmt,...) |
| apop_data * | apop_t_test (gsl_vector *a, gsl_vector *b) |
| int | apop_table_exists (char const *name, char remove) |
| double | apop_test (double statistic, char *distribution, double p1, double p2, char tail) |
| apop_data * | apop_test_anova_independence (apop_data *d) |
| apop_data * | apop_test_fisher_exact (apop_data *intab) |
| apop_data * | apop_test_kolmogorov (apop_model *m1, apop_model *m2) |
| apop_data * | apop_text_alloc (apop_data *in, const size_t row, const size_t col) |
| apop_data * | apop_text_fill_base (apop_data *data, char *text[]) |
| void | apop_text_free (char ***freeme, int rows, int cols) |
| char * | apop_text_paste (apop_data const *strings, char *between, char *before, char *after, char *between_cols, int(*prune)(apop_data *, int, int, void *), void *prune_parameter) |
| int | apop_text_set (apop_data *in, const size_t row, const size_t col, const char *fmt,...) |
| apop_data * | apop_text_to_data (char const *text_file, int has_row_names, int has_col_names, int const *field_ends, char const *delimiters) |
| int | apop_text_to_db (char const *text_file, char *tabname, int has_row_names, int has_col_names, char **field_names, int const *field_ends, apop_data *field_params, char *table_params, char const *delimiters, char if_table_exists) |
| apop_data * | apop_text_unique_elements (const apop_data *d, size_t col) |
| apop_model * | apop_update (apop_data *data, apop_model *prior, apop_model *likelihood, gsl_rng *rng) |
| void | apop_vector_apply (gsl_vector *v, void(*fn)(double *)) |
| int | apop_vector_bounded (const gsl_vector *in, long double max) |
| gsl_vector * | apop_vector_copy (const gsl_vector *in) |
| double | apop_vector_correlation (const gsl_vector *ina, const gsl_vector *inb, const gsl_vector *weights) |
| double | apop_vector_cov (gsl_vector const *v1, gsl_vector const *v2, gsl_vector const *weights) |
| double | apop_vector_distance (const gsl_vector *ina, const gsl_vector *inb, const char metric, const double norm) |
| long double | apop_vector_entropy (gsl_vector *in) |
| void | apop_vector_exp (gsl_vector *v) |
| gsl_vector * | apop_vector_fill_base (gsl_vector *in, double[]) |
| double | apop_vector_kurtosis (const gsl_vector *in) |
| double | apop_vector_kurtosis_pop (gsl_vector const *v, gsl_vector const *weights) |
| void | apop_vector_log (gsl_vector *v) |
| void | apop_vector_log10 (gsl_vector *v) |
| gsl_vector * | apop_vector_map (const gsl_vector *v, double(*fn)(double)) |
| double | apop_vector_map_sum (const gsl_vector *in, double(*fn)(double)) |
| double | apop_vector_mean (gsl_vector const *v, gsl_vector const *weights) |
| gsl_vector * | apop_vector_moving_average (gsl_vector *, size_t) |
| void | apop_vector_normalize (gsl_vector *in, gsl_vector **out, const char normalization_type) |
| double * | apop_vector_percentiles (gsl_vector *data, char rounding) |
| void | apop_vector_print (gsl_vector *data, Output_declares) |
| void | apop_vector_print (gsl_vector *data, char const *output_name, FILE *output_pipe, char output_type, char output_append) |
| gsl_vector * | apop_vector_realloc (gsl_vector *v, size_t newheight) |
| void | apop_vector_show (const gsl_vector *data) |
| double | apop_vector_skew (const gsl_vector *in) |
| double | apop_vector_skew_pop (gsl_vector const *v, gsl_vector const *weights) |
| gsl_vector * | apop_vector_stack (gsl_vector *v1, gsl_vector const *v2, char inplace) |
| long double | apop_vector_sum (const gsl_vector *in) |
| gsl_matrix * | apop_vector_to_matrix (const gsl_vector *in, char row_col) |
| gsl_vector * | apop_vector_unique_elements (const gsl_vector *v) |
| double | apop_vector_var (gsl_vector const *v, gsl_vector const *weights) |
| double | apop_vector_var_m (const gsl_vector *in, const double mean) |
Variables | |
| apop_model * | apop_bernoulli |
| apop_model * | apop_bernoulli |
| apop_model * | apop_bernoulli |
| apop_model * | apop_bernoulli |
| apop_model * | apop_beta |
| apop_model * | apop_beta |
| apop_model * | apop_beta |
| apop_model * | apop_beta |
| apop_model * | apop_binomial |
| apop_model * | apop_binomial |
| apop_model * | apop_binomial |
| apop_model * | apop_binomial |
| apop_model * | apop_chi_squared |
| apop_model * | apop_chi_squared |
| apop_model * | apop_chi_squared |
| apop_model * | apop_chi_squared |
| apop_model * | apop_composition |
| apop_model * | apop_composition |
| apop_model * | apop_composition |
| apop_model * | apop_composition |
| apop_model * | apop_coordinate_transform |
| apop_model * | apop_coordinate_transform |
| apop_model * | apop_coordinate_transform |
| apop_model * | apop_coordinate_transform |
| apop_model * | apop_cross |
| apop_model * | apop_cross |
| apop_model * | apop_cross |
| apop_model * | apop_cross |
| apop_model * | apop_dconstrain |
| apop_model * | apop_dconstrain |
| apop_model * | apop_dconstrain |
| apop_model * | apop_dconstrain |
| apop_model * | apop_dirichlet |
| apop_model * | apop_dirichlet |
| apop_model * | apop_dirichlet |
| apop_model * | apop_dirichlet |
| apop_model * | apop_exponential |
| apop_model * | apop_exponential |
| apop_model * | apop_exponential |
| apop_model * | apop_exponential |
| apop_model * | apop_f_distribution |
| apop_model * | apop_f_distribution |
| apop_model * | apop_f_distribution |
| apop_model * | apop_f_distribution |
| apop_model * | apop_gamma |
| apop_model * | apop_gamma |
| apop_model * | apop_gamma |
| apop_model * | apop_gamma |
| apop_model * | apop_improper_uniform |
| apop_model * | apop_improper_uniform |
| apop_model * | apop_improper_uniform |
| apop_model * | apop_improper_uniform |
| apop_model * | apop_iv |
| apop_model * | apop_iv |
| apop_model * | apop_iv |
| apop_model * | apop_iv |
| apop_model * | apop_kernel_density |
| apop_model * | apop_kernel_density |
| apop_model * | apop_kernel_density |
| apop_model * | apop_kernel_density |
| apop_model * | apop_loess |
| apop_model * | apop_loess |
| apop_model * | apop_loess |
| apop_model * | apop_loess |
| apop_model * | apop_logit |
| apop_model * | apop_logit |
| apop_model * | apop_logit |
| apop_model * | apop_logit |
| apop_model * | apop_lognormal |
| apop_model * | apop_lognormal |
| apop_model * | apop_lognormal |
| apop_model * | apop_lognormal |
| apop_model * | apop_mixture |
| apop_model * | apop_mixture |
| apop_model * | apop_mixture |
| apop_model * | apop_mixture |
| apop_model * | apop_multinomial |
| apop_model * | apop_multinomial |
| apop_model * | apop_multinomial |
| apop_model * | apop_multinomial |
| apop_model * | apop_multivariate_normal |
| apop_model * | apop_multivariate_normal |
| apop_model * | apop_multivariate_normal |
| apop_model * | apop_multivariate_normal |
| apop_model * | apop_normal |
| apop_model * | apop_normal |
| apop_model * | apop_normal |
| apop_model * | apop_normal |
| apop_model * | apop_ols |
| apop_model * | apop_ols |
| apop_model * | apop_ols |
| apop_model * | apop_ols |
| apop_opts_type | apop_opts |
| apop_opts_type | apop_opts |
| apop_opts_type | apop_opts |
| apop_opts_type | apop_opts |
| apop_opts_type | apop_opts |
| apop_opts_type | apop_opts |
| apop_model * | apop_pmf |
| apop_model * | apop_pmf |
| apop_model * | apop_pmf |
| apop_model * | apop_pmf |
| apop_model * | apop_poisson |
| apop_model * | apop_poisson |
| apop_model * | apop_poisson |
| apop_model * | apop_poisson |
| apop_model * | apop_probit |
| apop_model * | apop_probit |
| apop_model * | apop_probit |
| apop_model * | apop_probit |
| apop_model * | apop_t_distribution |
| apop_model * | apop_t_distribution |
| apop_model * | apop_t_distribution |
| apop_model * | apop_t_distribution |
| apop_model * | apop_uniform |
| apop_model * | apop_uniform |
| apop_model * | apop_uniform |
| apop_model * | apop_uniform |
| apop_model * | apop_wls |
| apop_model * | apop_wls |
| apop_model * | apop_wls |
| apop_model * | apop_wls |
| apop_model * | apop_yule |
| apop_model * | apop_yule |
| apop_model * | apop_yule |
| apop_model * | apop_yule |
| apop_model * | apop_zipf |
| apop_model * | apop_zipf |
| apop_model * | apop_zipf |
| apop_model * | apop_zipf |
| #define Apop_c | ( | d, | |
| col | |||
| ) |
A macro to generate a temporary one-column view of apop_data set d, pulling out only column col. After this call, outd will be a pointer to this temporary view, that you can use as you would any apop_data set.
| #define Apop_c | ( | d, | |
| col | |||
| ) |
A macro to generate a temporary one-column view of apop_data set d, pulling out only column col. After this call, outd will be a pointer to this temporary view, that you can use as you would any apop_data set.
| #define Apop_c | ( | d, | |
| col | |||
| ) |
A macro to generate a temporary one-column view of apop_data set d, pulling out only column col. After this call, outd will be a pointer to this temporary view, that you can use as you would any apop_data set.
| #define Apop_c | ( | d, | |
| col | |||
| ) |
A macro to generate a temporary one-column view of apop_data set d, pulling out only column col. After this call, outd will be a pointer to this temporary view, that you can use as you would any apop_data set.
| #define Apop_col_t | ( | d, | |
| colname, | |||
| outd | |||
| ) |
After this call, v will hold a view of the apop_data set m. The view will consist only of a gsl_vector view of the column of the apop_data set m with name col_name. Unlike Apop_c, the second argument is a column name, that I'll look up using apop_name_find, and the third is the name of the view to be generated.
| #define Apop_col_t | ( | d, | |
| colname, | |||
| outd | |||
| ) |
After this call, v will hold a view of the apop_data set m. The view will consist only of a gsl_vector view of the column of the apop_data set m with name col_name. Unlike Apop_c, the second argument is a column name, that I'll look up using apop_name_find, and the third is the name of the view to be generated.
| #define Apop_col_t | ( | d, | |
| colname, | |||
| outd | |||
| ) |
After this call, v will hold a view of the apop_data set m. The view will consist only of a gsl_vector view of the column of the apop_data set m with name col_name. Unlike Apop_c, the second argument is a column name, that I'll look up using apop_name_find, and the third is the name of the view to be generated.
| #define Apop_col_t | ( | d, | |
| colname, | |||
| outd | |||
| ) |
After this call, v will hold a view of the apop_data set m. The view will consist only of a gsl_vector view of the column of the apop_data set m with name col_name. Unlike Apop_c, the second argument is a column name, that I'll look up using apop_name_find, and the third is the name of the view to be generated.
| #define Apop_col_tv | ( | m, | |
| col, | |||
| v | |||
| ) |
After this call, v will hold a gsl_vector view of the apop_data set m. The view will consist only of the column with name col_name. Unlike Apop_cv, the second argument is a column name, that I'll look up using apop_name_find, and the third is the name of the view to be generated.
| #define Apop_col_tv | ( | m, | |
| col, | |||
| v | |||
| ) |
After this call, v will hold a gsl_vector view of the apop_data set m. The view will consist only of the column with name col_name. Unlike Apop_cv, the second argument is a column name, that I'll look up using apop_name_find, and the third is the name of the view to be generated.
| #define Apop_col_tv | ( | m, | |
| col, | |||
| v | |||
| ) |
After this call, v will hold a gsl_vector view of the apop_data set m. The view will consist only of the column with name col_name. Unlike Apop_cv, the second argument is a column name, that I'll look up using apop_name_find, and the third is the name of the view to be generated.
| #define Apop_col_tv | ( | m, | |
| col, | |||
| v | |||
| ) |
After this call, v will hold a gsl_vector view of the apop_data set m. The view will consist only of the column with name col_name. Unlike Apop_cv, the second argument is a column name, that I'll look up using apop_name_find, and the third is the name of the view to be generated.
| #define Apop_cs | ( | d, | |
| colnum, | |||
| len | |||
| ) |
A macro to generate a temporary view of apop_data set d including only certain columns, beginning at column col and having length len.
The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.
| #define Apop_cs | ( | d, | |
| colnum, | |||
| len | |||
| ) |
A macro to generate a temporary view of apop_data set d including only certain columns, beginning at column col and having length len.
The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.
| #define Apop_cs | ( | d, | |
| colnum, | |||
| len | |||
| ) |
A macro to generate a temporary view of apop_data set d including only certain columns, beginning at column col and having length len.
The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.
| #define Apop_cs | ( | d, | |
| colnum, | |||
| len | |||
| ) |
A macro to generate a temporary view of apop_data set d including only certain columns, beginning at column col and having length len.
The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.
| #define Apop_cv | ( | data_to_view, | |
| col | |||
| ) |
A macro to generate a temporary one-column view of the matrix in an apop_data set d, pulling out only column col. The view is a gsl_vector set.
As usual, column -1 is the vector element of the apop_data set.
The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.
| #define Apop_cv | ( | data_to_view, | |
| col | |||
| ) |
A macro to generate a temporary one-column view of the matrix in an apop_data set d, pulling out only column col. The view is a gsl_vector set.
As usual, column -1 is the vector element of the apop_data set.
The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.
| #define Apop_cv | ( | data_to_view, | |
| col | |||
| ) |
A macro to generate a temporary one-column view of the matrix in an apop_data set d, pulling out only column col. The view is a gsl_vector set.
As usual, column -1 is the vector element of the apop_data set.
The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.
| #define Apop_cv | ( | data_to_view, | |
| col | |||
| ) |
A macro to generate a temporary one-column view of the matrix in an apop_data set d, pulling out only column col. The view is a gsl_vector set.
As usual, column -1 is the vector element of the apop_data set.
The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.
| #define apop_data_add_names | ( | dataset, | |
| type, | |||
| ... | |||
| ) |
Add a list of names to a data set.
NULL, and call the base function: NULL marker, this has good odds of segfaulting. You may prefer to use a for loop that inserts each name in turn using apop_name_add.| #define apop_data_add_names | ( | dataset, | |
| type, | |||
| ... | |||
| ) |
Add a list of names to a data set.
NULL, and call the base function: NULL marker, this has good odds of segfaulting. You may prefer to use a for loop that inserts each name in turn using apop_name_add.| #define apop_data_add_names | ( | dataset, | |
| type, | |||
| ... | |||
| ) |
Add a list of names to a data set.
NULL, and call the base function: NULL marker, this has good odds of segfaulting. You may prefer to use a for loop that inserts each name in turn using apop_name_add.| #define apop_data_add_names | ( | dataset, | |
| type, | |||
| ... | |||
| ) |
Add a list of names to a data set.
NULL, and call the base function: NULL marker, this has good odds of segfaulting. You may prefer to use a for loop that inserts each name in turn using apop_name_add.| #define apop_data_free | ( | freeme | ) |
Free an apop_data structure.
free(), it is safe to send in a NULL pointer (in which case the function does nothing). more pointer is not NULL, I will free the pointed-to data set first. If you don't want to free data sets down the chain, set more=NULL before calling this. freeme to NULL when it's done, because there's nothing safe you can do with the freed location, and you can later safely test conditions like if (data) .... | #define apop_data_free | ( | freeme | ) |
Free an apop_data structure.
free(), it is safe to send in a NULL pointer (in which case the function does nothing). more pointer is not NULL, I will free the pointed-to data set first. If you don't want to free data sets down the chain, set more=NULL before calling this. freeme to NULL when it's done, because there's nothing safe you can do with the freed location, and you can later safely test conditions like if (data) .... | #define apop_data_free | ( | freeme | ) |
Free an apop_data structure.
free(), it is safe to send in a NULL pointer (in which case the function does nothing). more pointer is not NULL, I will free the pointed-to data set first. If you don't want to free data sets down the chain, set more=NULL before calling this. freeme to NULL when it's done, because there's nothing safe you can do with the freed location, and you can later safely test conditions like if (data) .... | #define apop_data_free | ( | freeme | ) |
Free an apop_data structure.
free(), it is safe to send in a NULL pointer (in which case the function does nothing). more pointer is not NULL, I will free the pointed-to data set first. If you don't want to free data sets down the chain, set more=NULL before calling this. freeme to NULL when it's done, because there's nothing safe you can do with the freed location, and you can later safely test conditions like if (data) .... | #define apop_gaussian |
Alias for the apop_normal distribution, qv.
| #define apop_gaussian |
Alias for the apop_normal distribution, qv.
| #define apop_gaussian |
Alias for the apop_normal distribution, qv.
| #define apop_gaussian |
Alias for the apop_normal distribution, qv.
| #define Apop_mcv | ( | matrix_to_view, | |
| col | |||
| ) |
Get a vector view of a single column of a gsl_matrix.
| matrix_to_vew | A gsl_matrix. |
| row | An integer giving the column to be viewed. |
gsl_vector view of the given column. The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.| #define Apop_mcv | ( | matrix_to_view, | |
| col | |||
| ) |
Get a vector view of a single column of a gsl_matrix.
| matrix_to_vew | A gsl_matrix. |
| row | An integer giving the column to be viewed. |
gsl_vector view of the given column. The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.| #define Apop_mcv | ( | matrix_to_view, | |
| col | |||
| ) |
Get a vector view of a single column of a gsl_matrix.
| matrix_to_vew | A gsl_matrix. |
| row | An integer giving the column to be viewed. |
gsl_vector view of the given column. The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.| #define Apop_mcv | ( | matrix_to_view, | |
| col | |||
| ) |
Get a vector view of a single column of a gsl_matrix.
| matrix_to_vew | A gsl_matrix. |
| row | An integer giving the column to be viewed. |
gsl_vector view of the given column. The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.| #define apop_model_copy_set | ( | model, | |
| type, | |||
| ... | |||
| ) |
Copy a model and add a settings group. Useful for models that require a settings group to function. See Apop_settings_add_group.
| #define apop_model_copy_set | ( | model, | |
| type, | |||
| ... | |||
| ) |
Copy a model and add a settings group. Useful for models that require a settings group to function. See Apop_settings_add_group.
| #define apop_model_copy_set | ( | model, | |
| type, | |||
| ... | |||
| ) |
Copy a model and add a settings group. Useful for models that require a settings group to function. See Apop_settings_add_group.
| #define apop_model_copy_set | ( | model, | |
| type, | |||
| ... | |||
| ) |
Copy a model and add a settings group. Useful for models that require a settings group to function. See Apop_settings_add_group.
| #define apop_model_cross | ( | ... | ) |
Generate a model consisting of the cross product of several independent models. The output apop_model is a copy of apop_cross; see that model's documentation for details.
apop_opts.verbose >= 2.| error=='n' | First model input is NULL. |
Examples:
| #define apop_model_cross | ( | ... | ) |
Generate a model consisting of the cross product of several independent models. The output apop_model is a copy of apop_cross; see that model's documentation for details.
apop_opts.verbose >= 2.| error=='n' | First model input is NULL. |
Examples:
| #define apop_model_cross | ( | ... | ) |
Generate a model consisting of the cross product of several independent models. The output apop_model is a copy of apop_cross; see that model's documentation for details.
apop_opts.verbose >= 2.| error=='n' | First model input is NULL. |
Examples:
| #define apop_model_cross | ( | ... | ) |
Generate a model consisting of the cross product of several independent models. The output apop_model is a copy of apop_cross; see that model's documentation for details.
apop_opts.verbose >= 2.| error=='n' | First model input is NULL. |
Examples:
| #define apop_model_mixture | ( | ... | ) |
Produce a model as a linear combination of other models. See the documentation for the apop_mixture model.
| ... | A list of models, either all parameterized or all unparameterized. See examples in the apop_mixture documentation. |
| #define apop_model_mixture | ( | ... | ) |
Produce a model as a linear combination of other models. See the documentation for the apop_mixture model.
| ... | A list of models, either all parameterized or all unparameterized. See examples in the apop_mixture documentation. |
| #define apop_model_mixture | ( | ... | ) |
Produce a model as a linear combination of other models. See the documentation for the apop_mixture model.
| ... | A list of models, either all parameterized or all unparameterized. See examples in the apop_mixture documentation. |
| #define apop_model_mixture | ( | ... | ) |
Produce a model as a linear combination of other models. See the documentation for the apop_mixture model.
| ... | A list of models, either all parameterized or all unparameterized. See examples in the apop_mixture documentation. |
| #define Apop_model_set_settings | ( | model, | |
| ... | |||
| ) |
This is the complement to apop_model_set_parameters, for those models that are set up by adding settings group, rather than filling in a list of parameters.
For example, the apop_kernel_density model is built by adding a apop_kernel_density_settings group. From the example on the apop_kernel_density page:
The name of the model and the settings group to be built must match, which is the case for many model transformations, including apop_dconstrain and apop_cross. If the names do not match, use apop_model_copy_set.
| #define Apop_model_set_settings | ( | model, | |
| ... | |||
| ) |
This is the complement to apop_model_set_parameters, for those models that are set up by adding settings group, rather than filling in a list of parameters.
For example, the apop_kernel_density model is built by adding a apop_kernel_density_settings group. From the example on the apop_kernel_density page:
The name of the model and the settings group to be built must match, which is the case for many model transformations, including apop_dconstrain and apop_cross. If the names do not match, use apop_model_copy_set.
| #define Apop_model_set_settings | ( | model, | |
| ... | |||
| ) |
This is the complement to apop_model_set_parameters, for those models that are set up by adding settings group, rather than filling in a list of parameters.
For example, the apop_kernel_density model is built by adding a apop_kernel_density_settings group. From the example on the apop_kernel_density page:
The name of the model and the settings group to be built must match, which is the case for many model transformations, including apop_dconstrain and apop_cross. If the names do not match, use apop_model_copy_set.
| #define Apop_model_set_settings | ( | model, | |
| ... | |||
| ) |
This is the complement to apop_model_set_parameters, for those models that are set up by adding settings group, rather than filling in a list of parameters.
For example, the apop_kernel_density model is built by adding a apop_kernel_density_settings group. From the example on the apop_kernel_density page:
The name of the model and the settings group to be built must match, which is the case for many model transformations, including apop_dconstrain and apop_cross. If the names do not match, use apop_model_copy_set.
| #define Apop_mrv | ( | matrix_to_view, | |
| row | |||
| ) |
Get a vector view of a single row of a gsl_matrix.
| matrix_to_vew | A gsl_matrix. |
| row | An integer giving the row to be viewed. |
gsl_vector view of the given row. The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.See apop_vector_correlation for an example of use.
| #define Apop_mrv | ( | matrix_to_view, | |
| row | |||
| ) |
Get a vector view of a single row of a gsl_matrix.
| matrix_to_vew | A gsl_matrix. |
| row | An integer giving the row to be viewed. |
gsl_vector view of the given row. The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.See apop_vector_correlation for an example of use.
| #define Apop_mrv | ( | matrix_to_view, | |
| row | |||
| ) |
Get a vector view of a single row of a gsl_matrix.
| matrix_to_vew | A gsl_matrix. |
| row | An integer giving the row to be viewed. |
gsl_vector view of the given row. The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.See apop_vector_correlation for an example of use.
| #define Apop_mrv | ( | matrix_to_view, | |
| row | |||
| ) |
Get a vector view of a single row of a gsl_matrix.
| matrix_to_vew | A gsl_matrix. |
| row | An integer giving the row to be viewed. |
gsl_vector view of the given row. The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.See apop_vector_correlation for an example of use.
| #define Apop_notify | ( | verbosity, | |
| ... | |||
| ) |
Notify the user of errors, warning, or debug info.
writes to apop_opts.log_file, which is a FILE handle. The default is stderr, but use fopen to attach to a file.
| verbosity | At what verbosity level should the user be warned? E.g., if level==2, then print iff apop_opts.verbosity >= 2. |
| ... | The message to write to the log (presuming the verbosity level is high enough). This can be a printf-style format with following arguments, e.g., apop_notify(0, "Beta is currently %g", beta). |
| #define Apop_notify | ( | verbosity, | |
| ... | |||
| ) |
Notify the user of errors, warning, or debug info.
writes to apop_opts.log_file, which is a FILE handle. The default is stderr, but use fopen to attach to a file.
| verbosity | At what verbosity level should the user be warned? E.g., if level==2, then print iff apop_opts.verbosity >= 2. |
| ... | The message to write to the log (presuming the verbosity level is high enough). This can be a printf-style format with following arguments, e.g., apop_notify(0, "Beta is currently %g", beta). |
| #define Apop_notify | ( | verbosity, | |
| ... | |||
| ) |
Notify the user of errors, warning, or debug info.
writes to apop_opts.log_file, which is a FILE handle. The default is stderr, but use fopen to attach to a file.
| verbosity | At what verbosity level should the user be warned? E.g., if level==2, then print iff apop_opts.verbosity >= 2. |
| ... | The message to write to the log (presuming the verbosity level is high enough). This can be a printf-style format with following arguments, e.g., apop_notify(0, "Beta is currently %g", beta). |
| #define Apop_notify | ( | verbosity, | |
| ... | |||
| ) |
Notify the user of errors, warning, or debug info.
writes to apop_opts.log_file, which is a FILE handle. The default is stderr, but use fopen to attach to a file.
| verbosity | At what verbosity level should the user be warned? E.g., if level==2, then print iff apop_opts.verbosity >= 2. |
| ... | The message to write to the log (presuming the verbosity level is high enough). This can be a printf-style format with following arguments, e.g., apop_notify(0, "Beta is currently %g", beta). |
| #define Apop_r | ( | d, | |
| rownum | |||
| ) |
A macro to generate a temporary one-row view of apop_data set d, pulling out only row row. The view is also an apop_data set, with names and other decorations.
The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.
| #define Apop_r | ( | d, | |
| rownum | |||
| ) |
A macro to generate a temporary one-row view of apop_data set d, pulling out only row row. The view is also an apop_data set, with names and other decorations.
The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.
| #define Apop_r | ( | d, | |
| rownum | |||
| ) |
A macro to generate a temporary one-row view of apop_data set d, pulling out only row row. The view is also an apop_data set, with names and other decorations.
The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.
| #define Apop_r | ( | d, | |
| rownum | |||
| ) |
A macro to generate a temporary one-row view of apop_data set d, pulling out only row row. The view is also an apop_data set, with names and other decorations.
The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.
| #define Apop_row_t | ( | d, | |
| rowname, | |||
| outd | |||
| ) |
After this call, v will hold an apop_data view of an apop_data set m. The view will consist only of the row with name row_name. Unlike Apop_r, the second argument is a row name, that I'll look up using apop_name_find, and the third is the name of the view to be generated.
| #define Apop_row_t | ( | d, | |
| rowname, | |||
| outd | |||
| ) |
After this call, v will hold an apop_data view of an apop_data set m. The view will consist only of the row with name row_name. Unlike Apop_r, the second argument is a row name, that I'll look up using apop_name_find, and the third is the name of the view to be generated.
| #define Apop_row_t | ( | d, | |
| rowname, | |||
| outd | |||
| ) |
After this call, v will hold an apop_data view of an apop_data set m. The view will consist only of the row with name row_name. Unlike Apop_r, the second argument is a row name, that I'll look up using apop_name_find, and the third is the name of the view to be generated.
| #define Apop_row_t | ( | d, | |
| rowname, | |||
| outd | |||
| ) |
After this call, v will hold an apop_data view of an apop_data set m. The view will consist only of the row with name row_name. Unlike Apop_r, the second argument is a row name, that I'll look up using apop_name_find, and the third is the name of the view to be generated.
| #define Apop_row_tv | ( | m, | |
| row, | |||
| v | |||
| ) |
After this call, v will hold a gsl_vector view of an apop_data set m. The view will consist only of the row with name row_name. Unlike Apop_rv, the second argument is a row name, that I'll look up using apop_name_find, and the third is the name of the view to be generated.
| #define Apop_row_tv | ( | m, | |
| row, | |||
| v | |||
| ) |
After this call, v will hold a gsl_vector view of an apop_data set m. The view will consist only of the row with name row_name. Unlike Apop_rv, the second argument is a row name, that I'll look up using apop_name_find, and the third is the name of the view to be generated.
| #define Apop_row_tv | ( | m, | |
| row, | |||
| v | |||
| ) |
After this call, v will hold a gsl_vector view of an apop_data set m. The view will consist only of the row with name row_name. Unlike Apop_rv, the second argument is a row name, that I'll look up using apop_name_find, and the third is the name of the view to be generated.
| #define Apop_row_tv | ( | m, | |
| row, | |||
| v | |||
| ) |
After this call, v will hold a gsl_vector view of an apop_data set m. The view will consist only of the row with name row_name. Unlike Apop_rv, the second argument is a row name, that I'll look up using apop_name_find, and the third is the name of the view to be generated.
| #define Apop_rs | ( | d, | |
| rownum, | |||
| len | |||
| ) |
A macro to generate a temporary view of apop_data set d pulling only certain rows, beginning at row row and having height len.
The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.
| #define Apop_rs | ( | d, | |
| rownum, | |||
| len | |||
| ) |
A macro to generate a temporary view of apop_data set d pulling only certain rows, beginning at row row and having height len.
The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.
| #define Apop_rs | ( | d, | |
| rownum, | |||
| len | |||
| ) |
A macro to generate a temporary view of apop_data set d pulling only certain rows, beginning at row row and having height len.
The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.
| #define Apop_rs | ( | d, | |
| rownum, | |||
| len | |||
| ) |
A macro to generate a temporary view of apop_data set d pulling only certain rows, beginning at row row and having height len.
The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.
| #define Apop_rv | ( | data_to_view, | |
| row | |||
| ) |
A macro to generate a temporary one-row view of the matrix in an apop_data set d, pulling out only row row. The view is a gsl_vector set.
The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.
| #define Apop_rv | ( | data_to_view, | |
| row | |||
| ) |
A macro to generate a temporary one-row view of the matrix in an apop_data set d, pulling out only row row. The view is a gsl_vector set.
The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.
| #define Apop_rv | ( | data_to_view, | |
| row | |||
| ) |
A macro to generate a temporary one-row view of the matrix in an apop_data set d, pulling out only row row. The view is a gsl_vector set.
The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.
| #define Apop_rv | ( | data_to_view, | |
| row | |||
| ) |
A macro to generate a temporary one-row view of the matrix in an apop_data set d, pulling out only row row. The view is a gsl_vector set.
The view is automatically allocated, and disappears as soon as the program leaves the scope in which it is declared.
| #define Apop_settings_add_group | ( | model, | |
| type, | |||
| ... | |||
| ) |
Add a settings group. The first two arguments (the model you are attaching to and the settings group name) are mandatory, and then you can use the Designated initializers syntax to specify default values (if any).
See Settings groups, Optimization, or Apop_settting_set for examples.
| #define Apop_settings_add_group | ( | model, | |
| type, | |||
| ... | |||
| ) |
Add a settings group. The first two arguments (the model you are attaching to and the settings group name) are mandatory, and then you can use the Designated initializers syntax to specify default values (if any).
See Settings groups, Optimization, or Apop_settting_set for examples.
| #define Apop_settings_add_group | ( | model, | |
| type, | |||
| ... | |||
| ) |
Add a settings group. The first two arguments (the model you are attaching to and the settings group name) are mandatory, and then you can use the Designated initializers syntax to specify default values (if any).
See Settings groups, Optimization, or Apop_settting_set for examples.
| #define Apop_settings_add_group | ( | model, | |
| type, | |||
| ... | |||
| ) |
Add a settings group. The first two arguments (the model you are attaching to and the settings group name) are mandatory, and then you can use the Designated initializers syntax to specify default values (if any).
See Settings groups, Optimization, or Apop_settting_set for examples.
| #define Apop_settings_copy | ( | name, | |
| ... | |||
| ) |
A convenience macro for declaring the copy function for a new settings group. See Writing new settings groups for details and an example.
| #define Apop_settings_copy | ( | name, | |
| ... | |||
| ) |
A convenience macro for declaring the copy function for a new settings group. See Writing new settings groups for details and an example.
| #define Apop_settings_copy | ( | name, | |
| ... | |||
| ) |
A convenience macro for declaring the copy function for a new settings group. See Writing new settings groups for details and an example.
| #define Apop_settings_copy | ( | name, | |
| ... | |||
| ) |
A convenience macro for declaring the copy function for a new settings group. See Writing new settings groups for details and an example.
| #define Apop_settings_declarations | ( | ysg | ) |
Put this in your header file to declare the init, copy, and free functions for ysg_settings. Of course, these functions will also have to be defined in a .c file using Apop_settings_init, Apop_settings_copy, and Apop_settings_free.
| #define Apop_settings_declarations | ( | ysg | ) |
Put this in your header file to declare the init, copy, and free functions for ysg_settings. Of course, these functions will also have to be defined in a .c file using Apop_settings_init, Apop_settings_copy, and Apop_settings_free.
| #define Apop_settings_declarations | ( | ysg | ) |
Put this in your header file to declare the init, copy, and free functions for ysg_settings. Of course, these functions will also have to be defined in a .c file using Apop_settings_init, Apop_settings_copy, and Apop_settings_free.
| #define Apop_settings_declarations | ( | ysg | ) |
Put this in your header file to declare the init, copy, and free functions for ysg_settings. Of course, these functions will also have to be defined in a .c file using Apop_settings_init, Apop_settings_copy, and Apop_settings_free.
| #define Apop_settings_free | ( | name, | |
| ... | |||
| ) |
A convenience macro for declaring the delete function for a new settings group. See Writing new settings groups for details and an example.
| #define Apop_settings_free | ( | name, | |
| ... | |||
| ) |
A convenience macro for declaring the delete function for a new settings group. See Writing new settings groups for details and an example.
| #define Apop_settings_free | ( | name, | |
| ... | |||
| ) |
A convenience macro for declaring the delete function for a new settings group. See Writing new settings groups for details and an example.
| #define Apop_settings_free | ( | name, | |
| ... | |||
| ) |
A convenience macro for declaring the delete function for a new settings group. See Writing new settings groups for details and an example.
| #define Apop_settings_get | ( | model, | |
| type, | |||
| setting | |||
| ) |
Retrieves a setting from a model. See Apop_settings_get_group to pull the entire group.
| model | An apop_model. |
| type | A string giving the type of the settings group you are retrieving, without the _settings ending. E.g., for an apop_mle_settings group, use apop_mle. |
| setting | The struct element you want to retrieve. |
| #define Apop_settings_get | ( | model, | |
| type, | |||
| setting | |||
| ) |
Retrieves a setting from a model. See Apop_settings_get_group to pull the entire group.
| model | An apop_model. |
| type | A string giving the type of the settings group you are retrieving, without the _settings ending. E.g., for an apop_mle_settings group, use apop_mle. |
| setting | The struct element you want to retrieve. |
| #define Apop_settings_get | ( | model, | |
| type, | |||
| setting | |||
| ) |
Retrieves a setting from a model. See Apop_settings_get_group to pull the entire group.
| model | An apop_model. |
| type | A string giving the type of the settings group you are retrieving, without the _settings ending. E.g., for an apop_mle_settings group, use apop_mle. |
| setting | The struct element you want to retrieve. |
| #define Apop_settings_get | ( | model, | |
| type, | |||
| setting | |||
| ) |
Retrieves a setting from a model. See Apop_settings_get_group to pull the entire group.
| model | An apop_model. |
| type | A string giving the type of the settings group you are retrieving, without the _settings ending. E.g., for an apop_mle_settings group, use apop_mle. |
| setting | The struct element you want to retrieve. |
| #define Apop_settings_get_group | ( | m, | |
| type | |||
| ) |
Retrieves a settings group from a model. See Apop_settings_get to just pull a single item from within the settings group.
This macro returns NULL if a group of type type_settings isn't found attached to model m, so you can easily put it in a conditional like
| m | An apop_model |
| type | A string giving the type of the settings group you are retrieving. E.g., for an apop_mle_settings group, use only apop_mle. |
NULL if not found). | #define Apop_settings_get_group | ( | m, | |
| type | |||
| ) |
Retrieves a settings group from a model. See Apop_settings_get to just pull a single item from within the settings group.
This macro returns NULL if a group of type type_settings isn't found attached to model m, so you can easily put it in a conditional like
| m | An apop_model |
| type | A string giving the type of the settings group you are retrieving. E.g., for an apop_mle_settings group, use only apop_mle. |
NULL if not found). | #define Apop_settings_get_group | ( | m, | |
| type | |||
| ) |
Retrieves a settings group from a model. See Apop_settings_get to just pull a single item from within the settings group.
This macro returns NULL if a group of type type_settings isn't found attached to model m, so you can easily put it in a conditional like
| m | An apop_model |
| type | A string giving the type of the settings group you are retrieving. E.g., for an apop_mle_settings group, use only apop_mle. |
NULL if not found). | #define Apop_settings_get_group | ( | m, | |
| type | |||
| ) |
Retrieves a settings group from a model. See Apop_settings_get to just pull a single item from within the settings group.
This macro returns NULL if a group of type type_settings isn't found attached to model m, so you can easily put it in a conditional like
| m | An apop_model |
| type | A string giving the type of the settings group you are retrieving. E.g., for an apop_mle_settings group, use only apop_mle. |
NULL if not found). | #define Apop_settings_init | ( | name, | |
| ... | |||
| ) |
A convenience macro for declaring the initialization function for a new settings group. See Writing new settings groups for details and an example.
| #define Apop_settings_init | ( | name, | |
| ... | |||
| ) |
A convenience macro for declaring the initialization function for a new settings group. See Writing new settings groups for details and an example.
| #define Apop_settings_init | ( | name, | |
| ... | |||
| ) |
A convenience macro for declaring the initialization function for a new settings group. See Writing new settings groups for details and an example.
| #define Apop_settings_init | ( | name, | |
| ... | |||
| ) |
A convenience macro for declaring the initialization function for a new settings group. See Writing new settings groups for details and an example.
| #define Apop_settings_rm_group | ( | m, | |
| type | |||
| ) |
Removes a settings group from a model's list.
| #define Apop_settings_rm_group | ( | m, | |
| type | |||
| ) |
Removes a settings group from a model's list.
| #define Apop_settings_rm_group | ( | m, | |
| type | |||
| ) |
Removes a settings group from a model's list.
| #define Apop_settings_rm_group | ( | m, | |
| type | |||
| ) |
Removes a settings group from a model's list.
| #define Apop_settings_set | ( | model, | |
| type, | |||
| setting, | |||
| data | |||
| ) |
Modifies a single element of a settings group to the given value.
For example,
model==NULL, fails silently. model!=NULL but the given settings group is not found attached to the model, set model->error='s'. | #define Apop_settings_set | ( | model, | |
| type, | |||
| setting, | |||
| data | |||
| ) |
Modifies a single element of a settings group to the given value.
For example,
model==NULL, fails silently. model!=NULL but the given settings group is not found attached to the model, set model->error='s'. | #define Apop_settings_set | ( | model, | |
| type, | |||
| setting, | |||
| data | |||
| ) |
Modifies a single element of a settings group to the given value.
For example,
model==NULL, fails silently. model!=NULL but the given settings group is not found attached to the model, set model->error='s'. | #define Apop_settings_set | ( | model, | |
| type, | |||
| setting, | |||
| data | |||
| ) |
Modifies a single element of a settings group to the given value.
For example,
model==NULL, fails silently. model!=NULL but the given settings group is not found attached to the model, set model->error='s'. | #define Apop_stopif | ( | test, | |
| onfail, | |||
| level, | |||
| ... | |||
| ) |
Execute an action and print a message to the current FILE handle held by apop_opts.log_file (default: stderr).
| test | The expression that, if true, triggers the action. |
| onfail | If the assertion fails, do this. E.g., out->error='x'; return GSL_NAN. Notice that it is OK to include several lines of semicolon-separated code here, but if you have a lot to do, the most readable option may be goto outro, plus an appropriately-labeled section at the end of your function. |
| level | Print the warning message only if apop_opts.verbose is greater than or equal to this. Zero usually works, but for minor infractions use one, or for more verbose debugging output use 2. |
| ... | The error message in printf form, plus any arguments to be inserted into the printf string. I'll provide the function name and a carriage return. |
Some examples:
apop_opts.stop_on_warning is nonzero and not 'v', then a failed test halts via abort(), even if the apop_opts.verbose level is set so that the warning message doesn't print to screen. Use this when running via debugger. apop_opts.stop_on_warning is 'v', then a failed test halts via abort() iff the verbosity level is high enough to print the error. | #define Apop_stopif | ( | test, | |
| onfail, | |||
| level, | |||
| ... | |||
| ) |
Execute an action and print a message to the current FILE handle held by apop_opts.log_file (default: stderr).
| test | The expression that, if true, triggers the action. |
| onfail | If the assertion fails, do this. E.g., out->error='x'; return GSL_NAN. Notice that it is OK to include several lines of semicolon-separated code here, but if you have a lot to do, the most readable option may be goto outro, plus an appropriately-labeled section at the end of your function. |
| level | Print the warning message only if apop_opts.verbose is greater than or equal to this. Zero usually works, but for minor infractions use one, or for more verbose debugging output use 2. |
| ... | The error message in printf form, plus any arguments to be inserted into the printf string. I'll provide the function name and a carriage return. |
Some examples:
apop_opts.stop_on_warning is nonzero and not 'v', then a failed test halts via abort(), even if the apop_opts.verbose level is set so that the warning message doesn't print to screen. Use this when running via debugger. apop_opts.stop_on_warning is 'v', then a failed test halts via abort() iff the verbosity level is high enough to print the error. | #define Apop_stopif | ( | test, | |
| onfail, | |||
| level, | |||
| ... | |||
| ) |
Execute an action and print a message to the current FILE handle held by apop_opts.log_file (default: stderr).
| test | The expression that, if true, triggers the action. |
| onfail | If the assertion fails, do this. E.g., out->error='x'; return GSL_NAN. Notice that it is OK to include several lines of semicolon-separated code here, but if you have a lot to do, the most readable option may be goto outro, plus an appropriately-labeled section at the end of your function. |
| level | Print the warning message only if apop_opts.verbose is greater than or equal to this. Zero usually works, but for minor infractions use one, or for more verbose debugging output use 2. |
| ... | The error message in printf form, plus any arguments to be inserted into the printf string. I'll provide the function name and a carriage return. |
Some examples:
apop_opts.stop_on_warning is nonzero and not 'v', then a failed test halts via abort(), even if the apop_opts.verbose level is set so that the warning message doesn't print to screen. Use this when running via debugger. apop_opts.stop_on_warning is 'v', then a failed test halts via abort() iff the verbosity level is high enough to print the error. | #define Apop_stopif | ( | test, | |
| onfail, | |||
| level, | |||
| ... | |||
| ) |
Execute an action and print a message to the current FILE handle held by apop_opts.log_file (default: stderr).
| test | The expression that, if true, triggers the action. |
| onfail | If the assertion fails, do this. E.g., out->error='x'; return GSL_NAN. Notice that it is OK to include several lines of semicolon-separated code here, but if you have a lot to do, the most readable option may be goto outro, plus an appropriately-labeled section at the end of your function. |
| level | Print the warning message only if apop_opts.verbose is greater than or equal to this. Zero usually works, but for minor infractions use one, or for more verbose debugging output use 2. |
| ... | The error message in printf form, plus any arguments to be inserted into the printf string. I'll provide the function name and a carriage return. |
Some examples:
apop_opts.stop_on_warning is nonzero and not 'v', then a failed test halts via abort(), even if the apop_opts.verbose level is set so that the warning message doesn't print to screen. Use this when running via debugger. apop_opts.stop_on_warning is 'v', then a failed test halts via abort() iff the verbosity level is high enough to print the error. | #define Apop_subm | ( | matrix_to_view, | |
| srow, | |||
| scol, | |||
| nrows, | |||
| ncols | |||
| ) |
Generate a view of a submatrix within a gsl_matrix. Like Apop_r, et al., the view is an automatically-allocated variable that is lost once the program flow leaves the scope in which it is declared.
| data_to_view | The root matrix |
| srow | the first row (in the root matrix) of the top of the submatrix |
| scol | the first column (in the root matrix) of the left edge of the submatrix |
| nrows | number of rows in the submatrix |
| ncols | number of columns in the submatrix |
gsl_matrix. | #define Apop_subm | ( | matrix_to_view, | |
| srow, | |||
| scol, | |||
| nrows, | |||
| ncols | |||
| ) |
Generate a view of a submatrix within a gsl_matrix. Like Apop_r, et al., the view is an automatically-allocated variable that is lost once the program flow leaves the scope in which it is declared.
| data_to_view | The root matrix |
| srow | the first row (in the root matrix) of the top of the submatrix |
| scol | the first column (in the root matrix) of the left edge of the submatrix |
| nrows | number of rows in the submatrix |
| ncols | number of columns in the submatrix |
gsl_matrix. | #define Apop_subm | ( | matrix_to_view, | |
| srow, | |||
| scol, | |||
| nrows, | |||
| ncols | |||
| ) |
Generate a view of a submatrix within a gsl_matrix. Like Apop_r, et al., the view is an automatically-allocated variable that is lost once the program flow leaves the scope in which it is declared.
| data_to_view | The root matrix |
| srow | the first row (in the root matrix) of the top of the submatrix |
| scol | the first column (in the root matrix) of the left edge of the submatrix |
| nrows | number of rows in the submatrix |
| ncols | number of columns in the submatrix |
gsl_matrix. | #define Apop_subm | ( | matrix_to_view, | |
| srow, | |||
| scol, | |||
| nrows, | |||
| ncols | |||
| ) |
Generate a view of a submatrix within a gsl_matrix. Like Apop_r, et al., the view is an automatically-allocated variable that is lost once the program flow leaves the scope in which it is declared.
| data_to_view | The root matrix |
| srow | the first row (in the root matrix) of the top of the submatrix |
| scol | the first column (in the root matrix) of the left edge of the submatrix |
| nrows | number of rows in the submatrix |
| ncols | number of columns in the submatrix |
gsl_matrix. | apop_data * apop_anova | ( | char * | table, |
| char * | data, | ||
| char * | grouping1, | ||
| char * | grouping2 | ||
| ) |
This function produces a traditional one- or two-way ANOVA table. It works from data in an SQL table, using queries of a form like select data from table group by grouping1, grouping2.
| table | The table to be queried. Anything that can go in an SQL from clause is OK, so this can be a plain table name or a temp table specification like (select ... ), with parens. |
| data | The name of the column holding the count or other such data |
| grouping1 | The name of the first column by which to group data |
| grouping2 | If this is NULL, then the function will return a one-way ANOVA. Otherwise, the name of the second column by which to group data in a two-way ANOVA. |
| int apop_arms_draw | ( | double * | out, |
| gsl_rng * | r, | ||
| apop_model * | m | ||
| ) |
Adaptive rejection Metropolis sampling, to make random draws from a univariate distribution.
The author, Wally Gilks, explains on http://www.amsta.leeds.ac.uk/~wally.gilks/adaptive.rejection/web_page/Welcome.html , that ``ARS works by constructing an envelope function of the log of the target density, which is then used in rejection sampling (see, for example, Ripley, 1987). Whenever a point is rejected by ARS, the envelope is updated to correspond more closely to the true log density, thereby reducing the chance of rejecting subsequent points. Fewer ARS rejection steps implies fewer point-evaluations of the log density.''
apop_arms_settings structure. The structure also holds a history of the points tested to date. That means that the system will be more accurate as more draws are made. It also means that if the parameters change, or you use apop_model_copy, you should call Apop_settings_rm_group(your_model, apop_arms) to clear the model of points that are not valid for a different situation. | gsl_vector * apop_array_to_vector | ( | double * | in, |
| int | size | ||
| ) |
Copies a one-dimensional array to a gsl_vector. The input array is undisturbed.
| in | An array of doubles. (No default. Must not be NULL); |
| size | How long line is. If this is zero or omitted, I'll guess using the sizeof(line)/sizeof(line[0]) trick, which will work for most arrays allocated using double [] and won't work for those allocated using double *. (default = auto-guess) |
gsl_vector, allocated and filled with a copy of (not a pointer to) the input data.NULL vector, you get a NULL pointer in return. I warn you of this if apop_opts.verbosity >=1 .| apop_model * apop_beta_from_mean_var | ( | double | m, |
| double | v | ||
| ) |
The Beta distribution is useful for modeling because it is bounded between zero and one, and can be either unimodal (if the variance is low) or bimodal (if the variance is high), and can have either a slant toward the bottom or top of the range (depending on the mean).
The distribution has two parameters, typically named 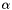 and
and  , which can be difficult to interpret. However, there is a one-to-one mapping between (alpha, beta) pairs and (mean, variance) pairs. Since we have good intuition about the meaning of means and variances, this function takes in a mean and variance, calculates alpha and beta behind the scenes, and returns the appropriate Beta distribution.
, which can be difficult to interpret. However, there is a one-to-one mapping between (alpha, beta) pairs and (mean, variance) pairs. Since we have good intuition about the meaning of means and variances, this function takes in a mean and variance, calculates alpha and beta behind the scenes, and returns the appropriate Beta distribution.
| m | The mean the Beta distribution should have. Notice that m is in [0,1]. |
| v | The variance which the Beta distribution should have. It is in (0, 1/12), where (1/12) is the variance of a Uniform(0,1) distribution. Funny things happen with variance near 1/12 and mean far from 1/2. |
apop_beta model and setting its parameters appropriately.| out->error=='r' | Range error: mean is not within [0, 1]. |
| apop_data * apop_bootstrap_cov | ( | apop_data * | data, |
| apop_model * | model, | ||
| gsl_rng * | rng, | ||
| int | iterations, | ||
| char | keep_boots, | ||
| char | ignore_nans, | ||
| apop_data ** | boot_store | ||
| ) |
Give me a data set and a model, and I'll give you the bootstrapped covariance matrix of the parameter estimates.
| data | The data set. An apop_data set where each row is a single data point. (No default) |
| model | An apop_model, whose estimate method will be used here. (No default) |
| iterations | How many bootstrap draws should I make? (default: 1,000) |
| rng | An RNG that you have initialized, probably with apop_rng_alloc. (Default: an RNG from apop_rng_get_thread) |
| boot_store | If not NULL, put the list of drawn parameter values here, with one parameter set per row. Sample use: The rows are packed via apop_data_pack, so use apop_data_unpack if needed. (Default: NULL) |
| ignore_nans | If 'y' and any of the elements in the estimation return NaN, then I will throw out that draw and try again. If 'n', then I will write that set of statistics to the list, NaN and all. I keep count of throw-aways; if there are more than iterations elements thrown out, then I throw an error and return with estimates using data I have so far. That is, I assume that NaNs are rare edge cases; if they are as common as good data, you might want to rethink how you are using the bootstrap mechanism. (Default: 'n') |
apop_data set whose matrix element is the estimated covariance matrix of the parameters. | out->error=='n' | NULL input data. |
| out->error=='N' | too many NaNs. |
This example is a sort of demonstration of the Central Limit Theorem. The model is a simulation, where each call to the estimation routine produces the mean/std dev of a set of draws from a Uniform Distribution. Because the simulation takes no inputs, apop_bootstrap_cov simply re-runs the simulation and calculates a sequence of mean/std dev pairs, and reports the covariance of that generated data set.
| double apop_cdf | ( | apop_data * | d, |
| apop_model * | m | ||
| ) |
Input a one-row data point/vector and a model; returns the area of the model's PDF beneath the given point.
By default, make random draws from the PDF and return the percentage of those draws beneath or equal to the given point. Many models have closed-form solutions that make no use of random draws.
See also apop_cdf_settings, which is the structure used to store draws already made (which means the second, third, ... calls to this function will take much less time than the first), the gsl_rng, and the number of draws to be made. These are handled without your involvement, but if you would like to change the number of draws from the default, add this group before calling apop_cdf :
Here are many examples using common, mostly symmetric distributions.
| void apop_crosstab_to_db | ( | apop_data * | in, |
| char * | tabname, | ||
| char * | row_col_name, | ||
| char * | col_col_name, | ||
| char * | data_col_name | ||
| ) |
See apop_db_to_crosstab for the storyline; this is the complement, which takes a crosstab and writes its values to the database.
For example, I would take
| c0 | c1 | |
| r0 | 2 | 3 |
| r1 | 0 | 4 |
and do the following writes to the database:
r0, r1, ... c0, c1, .... Text columns get their own names, t0, t1.| void apop_data_add_named_elmt | ( | apop_data * | d, |
| char * | name, | ||
| double | val | ||
| ) |
A convenience function to add a named element to a data set. Many of Apophenia's testing procedures use this to easily produce a column of named parameters. It is public as a convenience.
| d | The apop_data structure. Must not be NULL, but may be blank (as per allocation via apop_data_alloc ( ) ). |
| name | The name to add |
| val | the value to add to the set. |
NULL), I will call apop_vector_realloc internally to make space. Add a page to an apop_data set. It gets a name so you can find it later.
| dataset | The input data set, to which a page will be added. |
| newpage | The page to append |
| title | The name of the new page. |
NULL data set and apop_opts.verbose >=1 .| apop_data * apop_data_alloc | ( | const size_t | size1, |
| const size_t | size2, | ||
| const int | size3 | ||
| ) |
Allocate an apop_data structure.
apop_data_alloc(2,3,4): vector size, matrix rows, matrix cols. If the first argument is zero, you get a NULL vector. apop_data_alloc(2,3), would allocate just a matrix, leaving the vector NULL. apop_data_alloc(2), would allocate just a vector, leaving the matrix NULL. apop_data_alloc(), will produce a basically blank set, with out->matrix and out->vector set to NULL.For allocating the text part, see apop_text_alloc.
The weights vector is set to NULL. If you need it, allocate it via
| out->error=='a' | Allocation error. The matrix, vector, or names couldn't be malloced, which probably means that you requested a very large data set. |
NULL. But if even this much fails, your computer may be on fire and you should go put it out.| apop_data * apop_data_calloc | ( | const size_t | size1, |
| const size_t | size2, | ||
| const int | size3 | ||
| ) |
Allocate a apop_data structure, to be filled with data; set everything in the allocated portion to zero. See apop_data_alloc for details.
| out->error=='a' | allocation error; probably out of memory.
|
Copy one apop_data structure to another. That is, all data is duplicated.
Basically a front-end for apop_data_memcpy for those who prefer this sort of syntax.
If the data set has a more pointer, that will be followed and subsequent pages copied as well.
| in | the input data |
| out.error='a' | Allocation error. |
| out.error='c' | Cyclic link: D->more == D (may be later in the chain, e.g., D->more->more = D->more) You'll have only a partial copy. |
| out.error='d' | Dimension error; should never happen. |
| out.error='p' | Missing part error; should never happen. |
>=1. Returns the matrix of correlation coefficients  relating each column with each other.
relating each column with each other.
| in | A data matrix: rows are observations, columns are variables. If you give me a weights vector, I'll use it. |
| out->error='a' | Allocation error. |
Returns the sample variance/covariance matrix relating each column of the matrix to each other column.
| in | An apop_data set. If the weights vector is set, I'll take it into account. |
 , not
, not 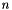 . If you need the population variance, use
. If you need the population variance, use | out->error='a' | Allocation error. |
| char apop_data_free_base | ( | apop_data * | freeme | ) |
Free the elements of the given apop_data set and then the apop_data set itself. Intended to be used by apop_data_free, a macro that calls this to free elements, then sets the value to NULL.
NULL. For typical cases, that's slightly more useful than this function.| freeme.error='c' | Circular linking is against the rules. If freeme->more == freeme, then I set freeme.error='c' and return. If you send in a structure like A -> B -> B, then both data sets A and B will be marked. |
0 on OK, 'c' on error. | double apop_data_get | ( | const apop_data * | data, |
| size_t | row, | ||
| int | col, | ||
| const char * | rowname, | ||
| const char * | colname, | ||
| const char * | page | ||
| ) |
Returns the data element at the given point.
In case of error (probably that you asked for a data point out of bounds), returns NAN. See the set/get page for details and examples.
| data | The data set. Must not be NULL. |
| row | The row number of the desired element. If rowname==NULL, default is zero. |
| col | The column number of the desired element. -1 indicates the vector. If colname==NULL, default is zero if the ->matrix element is not NULL and -1 if the ->matrix element is NULL and the ->vector element is not. |
| rowname | The row name of the desired element. If NULL, use the row number. |
| colname | The column name of the desired element. If NULL, use the column number. |
| page | The case-insensitive name of the page on which the element is found. If NULL, use first page. |
Factor names are stored in an auxiliary table with a name like "<categories for your_var>". Producing this name is annoying (and prevents us from eventually making it human-language independent), so use this function to get the list of factor names.
| data | The data set. (No default, must not be NULL) |
| col | The column in the main data set whose name I'll use to check for the factor name list. Vector==-1. (default=0) |
| type | If you are referring to a text column, use 't'. (default='d') |
It's good form to get a page from your data set by name, because you may not know the order for the pages, and the stepping through makes for dull code anyway (apop_data *page = dataset; while (page->more) page= page->more;).
| data | The apop_data set to use. No default; if NULL, gives a warning if apop_opts.verbose >=1 and returns NULL. |
| title | The name of the page to retrieve. Default="<Info>", which is the name of the page of additional estimation information returned by estimation routines (log likelihood, status, AIC, BIC, confidence intervals, ...). |
| match | If 'c', case-insensitive match (via strcasecmp); if 'e', exact match, if 'r' regular expression substring search (via apop_regex). Default='c'. |
NULL.If there is an NaN anywhere in the row of data (including the matrix, the vector, the weights, and the text) then delete the row from the data set.
NULL. apop_opts.nan_string is not NULL, then I will make case-insensitive comparisons to the text elements to check for bad data as well. inplace = 'y', then I'll free each element of the input data set and refill it with the pruned elements. I'll still take up (up to) twice the size of the data set in memory during the function. If every row has a NaN, then your apop_data set will end up with NULL vector, matrix, .... if inplace = 'n', then the original data set is left where it was, though internal elements may be moved. more element is ignored). | d | The data, with NaNs |
| inplace | If 'y', clear out the pointer-to-apop_data that you sent in and refill with the pruned data. If 'n', leave the set alone and return a new data set. Default='n'. |
inplace=='y', a pointer to the input, which was shortened in place. If the entire data set is cleared out, then this will be NULL. Copy one apop_data structure to another.
This function does not allocate the output structure or the vector, matrix, text, or weights elements—I assume you have already done this and got the dimensions right. I will assert that there is at least enough room in the destination for your data, and fail if the copy would write more elements than there are bins.
in and out have a more pointer, also copy subsequent page(s). | out | A structure that this function will fill. Must be preallocated with the appropriate sizes. |
| in | The input data. |
| out.error='d' | Dimension error. |
| out.error='p' | Part missing; e.g., in->matrix exists but out->matrix doesn't. |
| gsl_vector * apop_data_pack | ( | const apop_data * | in, |
| gsl_vector * | out, | ||
| char | more_pages, | ||
| char | use_info_pages | ||
| ) |
This function takes in an apop_data set and writes it as a single column of numbers, outputting a gsl_vector. It is valid to use the out_vector->data element as an array of doubles of size out_vector->data->size (i.e. its stride==1).
The complement is apop_data_unpack. I.e.,
will return the original data set (stripped of text and names).
| in | an apop_data set. No default; if NULL, return NULL. |
| out | If this is not NULL, then put the output here. The dimensions must match exactly. If NULL, then allocate a new data set. Default = NULL. |
| more_pages | If 'y', then follow the ->more pointer to fill subsequent pages; else fill only the first page. Informational pages will still be ignored, unless you set .use_info_pages='y' as well. Default = 'y'. |
| use_info_pages | Pages in XML-style brackets, such as <Covariance> will be ignored unless you set .use_info_pages='y'. Be sure that this is set to the same thing when you both pack and unpack. Default: 'n'. |
gsl_vector with the vector data (if any), then each row of data (if any), then the weights (if any), then the same for subsequent pages (if any && .more_pages=='y'). If out is not NULL, then this is out. | NULL | If you give me a vector as input, and its size is not correct, returns NULL. |
Say that you have added a long list of observations to a single apop_data set, meaning that each row has weight one. There are a huge number of duplicates, perhaps because there are a handful of types that keep repeating:
| Vector value | Text name | Weights |
| 12 | Dozen | 1 |
| 1 | Single | 1 |
| 2 | Pair | 1 |
| 2 | Pair | 1 |
| 1 | Single | 1 |
| 1 | Single | 1 |
| 2 | Pair | 1 |
| 2 | Pair | 1 |
Use this function to reduce this to a set of distinct values, with their weights adjusted accordingly:
| Vector value | Text name | Weights |
| 12 | Dozen | 1 |
| 1 | Single | 3 |
| 2 | Pair | 4 |
| in | An apop_data set that may have duplicate rows. As above, the data may be in text and/or numeric formats. |
weights vector, I will add those weights together as duplicates are merged. If there is no weights vector, I will create one, which is initially set to one for all values, and then aggregated as above. | void apop_data_print | ( | const apop_data * | data, |
| Output_declares | |||
| ) |
Print an apop_data set to a file, the database, or the screen, as determined by the .output_type.
Keep only the columns of a data set that you name. This is the function called internally by the apop_data_prune_columns macro. In most cases, you'll want to use that macro. An example of the two uses demonstrating the difference:
| d | The data set to prune. |
| colnames | A NULL-terminated list of names to retain. |
| double * apop_data_ptr | ( | apop_data * | data, |
| int | row, | ||
| int | col, | ||
| const char * | rowname, | ||
| const char * | colname, | ||
| const char * | page | ||
| ) |
Get a pointer to an element of an apop_data set.
NULL vector or matrix (as the case may be), or the row/column you requested is outside bounds, return NULL. | data | The data set. Must not be NULL. |
| row | The row number of the desired element. If rowname==NULL, default is zero. |
| col | The column number of the desired element. -1 indicates the vector. If colname==NULL, default is zero. |
| rowname | The row name of the desired element. If NULL, use the row number. |
| colname | The column name of the desired element. If NULL, use the column number. |
| page | The case-insensitive name of the page on which the element is found. If NULL, use first page. |
One often finds data where the column indicates the value of the data point. There may be two columns, and a mark in the first indicates a miss while a mark in the second is a hit. Or say that we have the following list of observations:
Then we could write this as:
because there are six 1s observed, four 2s observed, and two 3s observed. We call this rank format, because 1 (or zero) is typically the most common, 2 is second most common, et cetera.
This function takes in a list of observations, and aggregates them into a single row in rank format.
| in | The input apop_data set. If NULL, return NULL. |
| min_bins | If this is omitted, the number of bins is simply the largest number found. So if there are bins {0, 1, 2} and your data set happens to consist of 0 0 1 1 0, then I won't know to generate results with three bins where the last bin has a count of zero. Set .min_bins=2 to ensure that bin is included. |
The complement to this is apop_data_rank_compress; see that function's documentation for the story and an example.
This function takes in a data set where the zeroth column includes the count(s) of times that zero was observed, the first gives the count(s) of times that one was observed, et cetera. It outputs a data set whose vector element includes a list that has exactly the given frequency of zeros, ones, et cetera.
| void apop_data_rm_columns | ( | apop_data * | d, |
| int * | drop | ||
| ) |
Remove the columns of the apop_data set corresponding to a nonzero value in the drop vector.
| d | The apop_data structure to be pared down. |
| drop | An array of ints. If use[7]==1, then column seven will be cut from the output. A reminder: calloc(in->size2 , sizeof(int)) will fill your array with zeros on allocation, and memset(use, 1, in->size2 * sizeof(int)) will quickly fill an array of ints with nonzero values. apop_data_rm_rows |
Remove the first page from an apop_data set that matches a given name.
| data | The input data set, from which a page will be removed. No default. If NULL, maybe print a warning (see below). |
| title | The case-insensitive name of the page to remove. Default: "<Info>" |
| free_p | If 'y', then apop_data_free the page. Default: 'y'. |
apop_data page that I just pulled out. Thus, you can use this to pull a single page from a data set. I set that page's more pointer to NULL, to minimize any confusion about more-than-linear linked list topologies. If free_p=='y' (the default) or the page is not found, return NULL.->more pointer in the apop_data set is not to fully implement a linked list, but primarily to allow you to staple auxiliary information to a main data set. apop_opts.verbose >= 1 , print a warning. If the page is to be freed and apop_opts.verbose >= 2 , print a warning.more pointers in the apop_data set are adjusted accordingly. | apop_data * apop_data_rm_rows | ( | apop_data * | in, |
| int * | drop, | ||
| apop_fn_ir | do_drop, | ||
| void * | drop_parameter | ||
| ) |
Remove the rows set to one in the drop vector or for which the do_drop function returns one.
| in | the apop_data structure to be pared down |
| drop | a vector with as many elements as the max of the vector, matrix, or text parts of in, with a one marking those rows to be removed. |
| do_drop | A function that returns one for rows to drop and zero for rows to not drop. A sample function: apop_data_rm_rows will use Apop_r to get a subview of the input data set of height one, and send that subview to this function (and since arguments typically default to zero, you don't have to write out things like apop_data_get (onerow, .row=0, .col=0), which can help to keep things readable). |
| drop_parameter | If your do_drop function requires additional input, put it here and it will be passed through. |
NULL vector, matrix, weight, and text. Therefore, you may wish to check for NULL elements after use. I remove rownames, but leave the other names, in case you want to add new data rows. NULL, I return without doing anything, and print a warning if apop_opts.verbose >=2. If you provide both, I will drop the row if either the vector has a one in that row's position, or if the function returns a nonzero value. | int apop_data_set | ( | apop_data * | data, |
| size_t | row, | ||
| int | col, | ||
| const double | val, | ||
| const char * | colname, | ||
| const char * | rowname, | ||
| const char * | page | ||
| ) |
Set a data element. See the set/get page for details and examples.
_Thread_local keyword) or a version of GCC with the __thread extension enabled.gsl_vector_set(your_data->weights, row, val);. | data | The data set. Must not be NULL. |
| row | The row number of the desired element. If rowname==NULL, default is zero. |
| col | The column number of the desired element. -1 indicates the vector. If colname==NULL, default is zero. |
| rowname | The row name of the desired element. If NULL, use the row number. |
| colname | The column name of the desired element. If NULL, use the column number. |
| page | The case-insensitive name of the page on which the element is found. If NULL, use first page. |
| val | The value to give the point. |
| apop_data * apop_data_sort | ( | apop_data * | data, |
| apop_data * | sort_order, | ||
| char | asc, | ||
| char | inplace, | ||
| double * | col_order | ||
| ) |
Sort an apop_data set on an arbitrary sequence of columns.
The sort_order set is a one-row data set that should look like the data set being sorted. The easiest way to generate it is to use Apop_r to pull one row of the table, then copy and fill it. For each column you want used in the sort, assign a ranking giving whether the column should be sorted first, second, .... Columns you don't want used in the sorting should be set to NAN. Ties are broken by the earlier element in the default order (see below).
E.g., to sort by the last column of a five-column matrix first, then the next-to-last column, then the next-to-next-to-last, then by the first text column, then by the second text column:
To determine which columns are sorted at which step, I use only comparisons, not the actual numeric values. For example, (1, 2, 3) and (-1.32, 0, 27) work identically. For text, I use atof to convert the your text to a number, as in the example above that set text values of "4" and "5". A blank string, NaN numeric value, or NULL element in the apop_data set means that column will not be sorted.
strcasecmp. [exercise for the reader: modify the source to use Glib's locale-correct string sorting.]| data | The data set to be sorted. If NULL, this function is a no-op that returns NULL. |
| sort_order | An apop_data set describing the order in which columns are used for sorting, as above. If NULL, then sort by the vector, then each matrix column, then text, then weights, then row names. |
| inplace | If 'n', make a copy, else sort in place. (default: 'y'). |
| asc | If 'a', ascending; if 'd', descending. This is applied to all columns; column-by-column application is to do. (default: 'a'). |
| col_order | For internal use only. In your call, it should be NULL; you can leave this off your function call entirely and the Designated initializers syntax will takes care of it for you. |
inplace=='y' (the default), then this is the same as the input set.A few examples:
Split one input apop_data structure into two.
For the opposite operation, see apop_data_stack.
| in | The apop_data structure to split |
| splitpoint | The index of what will be the first row/column of the second data set. E.g., if this is -1 and r_or_c=='c', then the whole data set will be in the second data set; if this is the length of the matrix then the whole data set will be in the first data set. Another way to put it is that for values between zero and the matrix's size, splitpoint will equal the number of rows/columns in the first matrix. |
| r_or_c | If this is 'r' or 'R', then put some rows in the first data set and some in the second; of 'c' or 'C', split columns into first and second data sets. |
NULL pointer will be returned in that position. For example, for a data set of 50 rows, apop_data **out = apop_data_split(data, 100, 'r') sets out[0] = apop_data_copy(data) and out[1] = NULL.more pointer is ignored. apop_data->vector is taken to be the -1st element of the matrix. apop_data_free(in) after this. Put the first data set either on top of or to the left of the second data set.
For the opposite operation, see apop_data_split.
| m1 | the upper/rightmost data set (default = NULL) |
| m2 | the second data set (default = NULL) |
| posn | If 'r', stack rows of m1 above rows of m2 if 'c', stack columns of m1 to left of m2's (default = 'r') |
| inplace | If 'y', use apop_matrix_realloc and apop_vector_realloc to modify m1 in place. Otherwise, allocate a new apop_data set, leaving m1 undisturbed. (default='n') |
m1 | out->error=='a' | Allocation error. |
| out->error=='d' | Dimension error; couldn't make a complete copy. |
NULL, returns a copy of the other element, and if both are NULL, returns NULL. If m2 is NULL and inplace is 'y', returns the original m1 pointer unmodified. more is ignored. m1 and m2->vector doesn't appear in the output at all. m1, with the names for m2 appended to the row or column list, as appropriate. Put summary information about the columns of a table (mean, std dev, variance, min, median, max) in a table.
| indata | The table to be summarized. An apop_data structure. May have a weights element. |
| out->error='a' | Allocation error. |
| apop_data * apop_data_to_bins | ( | apop_data const * | indata, |
| apop_data const * | binspec, | ||
| int | bin_count, | ||
| char | close_top_bin | ||
| ) |
Create a histogram from data by putting data into bins of fixed width. Your input apop_data set may be multidimensional, and may include both vector and matrix parts, and the bins output will have corresponding dimension.
| indata | The input data that will be binned, one observation per row. This is copied and the copy will be modified. (No default) |
| binspec | This is an apop_data set with the same number of columns as indata. If you want a fixed size for the bins, then the first row of the bin spec is the bin width for each column. This allows you to specify a width for each dimension, or specify the same size for all with something like: The presumption is that the first bin starts at zero in all cases. You can add a second row to the spec to give the offset for each dimension. (default: NULL) |
| bin_count | If you don't provide a bin spec, I'll provide this many evenly-sized bins to cover the data set. (Default:  ) ) |
| close_top_bin | Normally, a bin covers the range from the point equal to its minimum to points strictly less than the minimum plus the width. if 'y', then the top bin includes points less than or equal to the upper bound. This solves the problem of displaying histograms where the top bin is just one point. (default: 'y' if binspec==NULL, else 'n') |
 bins to cover the range to the max element.
bins to cover the range to the max element. more pointer, if any, is not followed. NULL input, return NULL output. Print a warning if apop_opts.verbose >= 2.Iff you didn't give me a binspec, then I attach one to the output set as a page named <binspec>. This means that you can snap a second data set to the same grid using
Here is a sample program highlighting apop_data_to_bins and apop_data_pmf_compress .
| apop_data * apop_data_to_dummies | ( | apop_data * | d, |
| int | col, | ||
| char | type, | ||
| int | keep_first, | ||
| char | append, | ||
| char | remove | ||
| ) |
A utility to make a matrix of dummy variables. You give me a single vector that lists the category number for each item, and I'll produce a matrix with a single one in each row in the column specified.
After that, you have to decide what to do with the new matrix and the original data column.
.remove='y' option specifies that I should use apop_data_rm_columns to remove the column used to generate the dummies. Implemented only for type=='d'..append='y' or .append='e' I will run the above two lines for you. Your apop_data pointer will not change, but its matrix element will be reallocated (via apop_data_stack)..append='i', I will place the matrix of dummies in place, immediately after the data column you had specified. You will probably use this with .remove='y' to replace the single column with the new set of dummy columns. Bear in mind that if there are two or more dummy columns, adding columns will change subsequent column numbers; use apop_name_find to find columns instead of giving an explicit column number..append='i' and you asked for a text column, I will append to the end of the table, which is equivalent to append='e'.| d | The data set with the column to be dummified (No default.) |
| col | The column number to be transformed; -1==vector (default = 0) |
| type | 'd'==data column, 't'==text column. (default = 't') |
| keep_first | If 'n', return a matrix where each row has a one in the (column specified minus one). That is, the zeroth category is dropped, the first category has an entry in column zero, et cetera. If you don't know why this is useful, then this is what you need. If you know what you're doing and need something special, set this to 'y' and the first category won't be dropped. (default = 'n') |
| append | If 'e' or 'y', append the dummy grid to the end of the original data matrix. If 'i', insert in place, immediately after the original data column. (default = 'n') |
| remove | If 'y', remove the original data or text column. (default = 'n') |
matrix element is the one-zero matrix of dummies. If you used .append, then this is the main matrix. Also, I add a page named "\<categories for your_var\>" giving a reference table of names and column numbers (where your_var is the appropriate column heading). | out->error=='a' | allocation error |
| out->error=='d' | dimension error |
Convert a column of text or numbers into a column of numeric factors, which you can use for a multinomial probit/logit, for example.
If you don't run this on your data first, apop_probit and apop_logit default to running it on the vector or (if no vector) zeroth column of the matrix of the input apop_data set, because those models need a list of the unique values of the dependent variable.
| data | The data set to be modified in place. (No default. If NULL, returns NULL and a warning) |
| intype | If 't', then incol refers to text, if 'd', refers to the vector or matrix. (default = 't') |
| incol | The column in the text that will be converted. -1 is the vector. (default = 0) |
| outcol | The column in the data set where the numeric factors will be written (-1 means the vector). (default = 0) |
For example:
Notice that the query pulled a column of zeros for the sake of saving room for the factors. It reads column zero of the text, and writes it to column zero of the matrix.
Another example:
Here, the type column is converted to sequential integer factors and those factors overwrite the original data. Since a reference table is added as a second page of the apop_data set, you can recover the original values as needed.
apop_data set with only one column of text. Also, I add a page named "<categories for your_var>" giving a reference table of names and column numbers (where your_var is the appropriate column heading) use apop_data_get_factor_names to retrieve that table.| out->error=='a' | allocation error. |
| out->error=='d' | dimension error. |
NULL, I will allocate it for you. Transpose the matrix and text elements of the input data set, including the row/column names.
The vector and weights elements of the input data set are completely ignored (but see also apop_vector_to_matrix, which can convert a vector to a 1 X N matrix.) If copying, these other elements won't be present; if .inplace='y', it is up to you to handle these not-transposed elements correctly.
| in | The input apop_data set. If NULL, I return NULL. (default: NULL) |
| transpose_text | If 'y', then also transpose the text element. (default: 'y') |
| inplace | If 'y', transpose the input in place; if 'n', produce a transposed copy, leaving the original untouched. Due to how gsl_matrix_transpose_memcpy works, a copy will still be made, then copied to the original location. (default: 'y') |
inplace=='n', a newly alloced apop_data set, with the appropriately transposed matrix and/or text. The vector and weights elements will be NULL. If transpose_text='n', then the text element of the output set will also be NULL.inplace=='y', a pointer to the original data set, with matrix and (if transpose_text='y', text) transposed and vector and weights left in place untouched.gsl_matrix with no names or text, you may prefer to use gsl_matrix_transpose_memcpy. | void apop_data_unpack | ( | const gsl_vector * | in, |
| apop_data * | d, | ||
| char | use_info_pages | ||
| ) |
This is the complement to apop_data_pack, qv. It writes the gsl_vector produced by that function back to the apop_data set you provide. It overwrites the data in the vector and matrix elements and, if present, the weights (and that's it, so names or text are as before).
| in | A gsl_vector of the form produced by apop_data_pack. No default; must not be NULL. |
| d | That data set to be filled. Must be allocated to the correct size. No default; must not be NULL. |
| use_info_pages | Pages in XML-style brackets, such as <Covariance> will be ignored unless you set .use_info_pages='y'. Be sure that this is set to the same thing when you both pack and unpack. (Default: 'n'). |
apop_data set and have more entries in the vector to unpack, and the data to fill has a more element, then I will continue into subsequent pages. | int apop_db_close | ( | char | vacuum | ) |
Closes the database on disk. If you opened the database with apop_db_open(NULL), then this is basically optional.
| vacuum | 'v': vacuum—do clean-up to minimize the size of the database on disk. 'q': Don't bother; just close the database. (default = 'q') |
| int apop_db_open | ( | char const * | filename | ) |
If you want to use a database on the hard drive instead of memory, then call this once and only once before using any other database utilities.
With SQLite, if you want a disposable database which you won't use after the program ends, don't bother with this function.
The trade-offs between an on-disk database and an in-memory db are as one would expect: memory is faster, but the database is destroyed when the program exits.
MySQL users: either set the environment variable APOP_DB_ENGINE=mysql or set apop_opts.db_engine = 'm'.
The Apophenia package assumes you are only using a single database at a time. You can use the SQL attach function to load other databases, or see this blog post for further suggestions and sample code.
When you are done doing your database manipulations, call apop_db_close if writing to disk.
| filename | The name of a file on the hard drive on which to store the database. If NULL, then the database will be kept in memory (in which case, the other database functions will call this function for you and you don't need to bother). |
| apop_data * apop_db_to_crosstab | ( | char const * | tabname, |
| char const * | row, | ||
| char const * | col, | ||
| char const * | data, | ||
| char | is_aggregate | ||
| ) |
Give the name of a table in the database, and optional names of three of its columns: the x-dimension, the y-dimension, and the data. The output is a 2D matrix with rows indexed by 'row' and cols by 'col' and the cells filled with the entry in the 'data' column.
| tabname | The database table I'm querying. Anything that will work inside a from clause is OK, such as a subquery in parens. (no default; must not be NULL) |
| row | The column of the data set that will indicate the rows of the output crosstab (no default; must not be NULL) |
| col | The column of the data set that will indicate the columns of the output crosstab (no default; must not be NULL) |
| data | The column of the data set holding the data for the cells of the crosstab (default: count(*)) |
| is_aggregate | Set to 'y' if the data is a function like count(*) or sum(col). That is, set to 'y' if querying this would require a group by clause. (default: if I find an end-paren in datacol, 'y'; else 'n'.) |
NULL data set and if apop_opts.verbosity >= 1 print a warning.| out->error='n' | Name not found error. |
| out->error='q' | Query returned an empty table (which might mean that it just failed). |
apop_db_to_crosstab("datatab", "r1", "r2"). apop_db_to_crosstab that calls this function. | double apop_det_and_inv | ( | const gsl_matrix * | in, |
| gsl_matrix ** | out, | ||
| int | calc_det, | ||
| int | calc_inv | ||
| ) |
Calculate the determinant of a matrix, its inverse, or both, via LU decomposition. The in matrix is not destroyed in the process.
| in | The matrix to be inverted/determined. |
| out | If you want an inverse, this is where to place the matrix to be filled with the inverse. Will be allocated by the function. |
| calc_det | 0: Do not calculate the determinant. 1: Do. |
| calc_inv | 0: Do not calculate the inverse. 1: Do. |
calc_det == 1, then return the determinant. Otherwise, just returns zero. If calc_inv!=0, then *out is pointed to the matrix inverse. In case of difficulty, I will set *out=NULL and return NaN. A convenience function for dot products, which requires less prep and typing than the gsl_cblas_dgexx functions.
It makes use of the semi-overloading of the apop_data structure. d1 may be a vector or a matrix, and the same for d2, so this function can do vector dot matrix, matrix dot matrix, and so on. If d1 includes both a vector and a matrix, then later parameters will indicate which to use.
| d1 | the left part of  |
| d2 | the right part of  |
| form1 | 't' or 'p': transpose or prime d1->matrix, or, if d1->matrix is NULL, read d1->vector as a row vector.'n' or 0: use matrix if present; no transpose. (the default) 'v': ignore the matrix and use the vector. |
| form2 | As above, with d2. |
NULL and the matrix has the dot product; if either or both are vectors, the vector has the output and the matrix is NULL.| out->error='a' | Allocation error. |
| out->error='d' | dimension-matching error. |
| out->error='m' | GSL math error. |
| NULL | If you ask me to take the dot product of NULL, I return NULL. |
 and a
and a  matrix, and the system assumes that you meant to transpose the second, producing a
matrix, and the system assumes that you meant to transpose the second, producing a  output. Apophenia does not do this. First, it's ambiguous whether the output should be
output. Apophenia does not do this. First, it's ambiguous whether the output should be  or
or  . Second, your next run might have three observations, and two
. Second, your next run might have three observations, and two  matrices don't require transposition; auto-transposition thus creates situations where bugs can pop up on only some iterations of a loop.
matrices don't require transposition; auto-transposition thus creates situations where bugs can pop up on only some iterations of a loop.  a matrix, the vector is always treated as a row vector, meaning that a
a matrix, the vector is always treated as a row vector, meaning that a 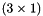 dot a
dot a 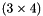 matrix is correct, and produces a
matrix is correct, and produces a  vector. For a matrix
vector. For a matrix  a vector, the vector is always treated as a column vector. Requests for transposing the vector are ignored in both cases.
a vector, the vector is always treated as a column vector. Requests for transposing the vector are ignored in both cases. 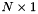 vector
vector  a
a  vector to produce an
vector to produce an  matrix, then use apop_vector_to_matrix to turn your vectors into matrices; see the example.
matrix, then use apop_vector_to_matrix to turn your vectors into matrices; see the example. d1 is a vector and d2 a matrix, then apop_dot(d1,d2,'t') won't work, because 't' now refers to d1. Instead use apop_dot(d1,d2,.form2='t') or apop_dot(d1,d2,0, 't') Sample code:
| int apop_draw | ( | double * | out, |
| gsl_rng * | r, | ||
| apop_model * | m | ||
| ) |
Draw from a model.
| out | An already-allocated array of doubles to be filled by the draw method. It must have size m->dsize. |
| r | A gsl_rng, probably allocated via apop_rng_alloc. Optional; if NULL, then I will call apop_rng_get_thread for an RNG. |
| m | The model from which to make draws. |
draw method, then this function will call it. out[0] is probably NAN on failure. | apop_model * apop_estimate | ( | apop_data * | d, |
| apop_model * | m | ||
| ) |
Estimate the parameters of a model given data.
This function copies the input model, preps it (see apop_prep), and calls m.estimate(d, m) (which users are encouraged to never call directly). If your model has no estimate method, then call apop_maximum_likelihood(d, m), with the default MLE settings.
| d | The data |
| m | The model |
parameters element filled in. | apop_data * apop_estimate_coefficient_of_determination | ( | apop_model * | m | ) |
Also known as 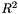 . Let
. Let  be the dependent variable,
be the dependent variable, 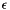 the residual,
the residual, 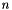 the number of data points, and
the number of data points, and 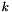 the number of independent vars (including the constant). Returns an apop_data set with the following entries (in the vector element):
the number of independent vars (including the constant). Returns an apop_data set with the following entries (in the vector element):


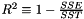

Internally allocates (and frees) a vector the size of your data set.
 apop_data table with the following fields:
apop_data table with the following fields: sss, use apop_data_get(sss, .rowname="SSE") to get the SSE, and so on for the other items.| m | A model. I use the pointer to the data set used for estimation and the info page named "<Predicted>". The Predicted page should include observed, expected, and residual columns, which I use to generate the sums of squared errors and residuals, et cetera. All generalized linear models produce a page with this name and of this form, as do a host of other models. Nothing keeps you from finding the 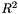 of, say, a kernel smooth; it is up to you to determine whether such a thing is appropriate to your given models and situation. of, say, a kernel smooth; it is up to you to determine whether such a thing is appropriate to your given models and situation. |
apop_estimate(yourdata, apop_ols) does this automatically "<Predicted>" page, print an error (iff apop_opts.verbose >=0) and return NULL. 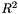 , is from the number of columns in the main data set's matrix (i.e. the first page; i.e. the set of parameters if this is the
, is from the number of columns in the main data set's matrix (i.e. the first page; i.e. the set of parameters if this is the parameters output from a model estimation). weights vector, I will find weighted SSE, SST, and SSR (and calculate the 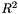 s using those values).
s using those values). | apop_model * apop_estimate_restart | ( | apop_model * | e, |
| apop_model * | copy, | ||
| char * | starting_pt, | ||
| double | boundary | ||
| ) |
Maximum likelihod searches are not guaranteed to find a global optimum, and it can be difficult to tune a search such that it covers a wide space, but also accurately hones in on the optimum. In both cases, one could restart the search using a different starting point or different parameters.
The simplest use of this function is to restart a model at the latest parameter estimates.
By adding a line to reduce the tolerance each round [e.g., Apop_settings_set(m, apop_mle, tolerance, pow(10,-i))], you can start broad and hone in on a precise optimum.
You may have a new estimation method, such as first doing a coarse simulated annealing search, then a fine conjugate gradient search. When reading this example, recall that the form for adding a new settings group differs from the form for modifying existing settings:
Only one estimate is returned, either the one you sent in or a new one. The loser (which may be the one you sent in) is freed, to prevent memory leaks.
| e | An apop_model that is the output from a prior MLE estimation. (No default, must not be NULL.) |
| copy | Another not-yet-parametrized model that will be re-estimated with (1) the same data and (2) a starting_pt as per the next setting (probably to the parameters of e). If this is NULL, then copy e. (Default = NULL) |
| starting_pt | "ep"=last estimate of the first model (i.e., its current parameter estimates) "es"= starting point originally used by the first model "np"=current parameters of the new (second) model "ns"=starting point specified by the new model's MLE settings. (default = "ep") |
| boundary | I test whether the starting point you give me has magintude greater than this bound, so I can warn you if there's divergence in your sequence of re-estimations. (default: 1e8) |
| apop_data * apop_f_test | ( | apop_model * | est, |
| apop_data * | contrast | ||
| ) |
Runs an F-test specified by q and c. See the chapter on hypothesis testing in Modeling With Data, p 309, which will tell you that:
![\[{N-K\over q} {({\bf Q}'\hat\beta - {\bf c})' [{\bf Q}' ({\bf X}'{\bf X})^{-1} {\bf Q}]^{-1} ({\bf Q}' \hat\beta - {\bf c}) \over {\bf u}' {\bf u} } \sim F_{q,N-K},\]](form_93.png)
and that's what this function is based on.
| est | An apop_model that you have already calculated. (No default) |
| contrast | An apop_data set whose matrix represents  and whose vector represents and whose vector represents  . Each row represents a hypothesis. (Defaults: if matrix is . Each row represents a hypothesis. (Defaults: if matrix is NULL, it is set to the identity matrix with the top row missing. If the vector is NULL, it is set to a zero matrix of length equal to the height of the contrast matrix. Thus, if the entire apop_data set is NULL or omitted, we are testing the hypothesis that all but 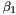 are zero.) are zero.) |
apop_data set with a few variants on the confidence with which we can reject the joint hypothesis. | out->error='a' | Allocation error. |
| out->error='d' | dimension-matching error. |
| out->error='i' | matrix inversion error. |
| out->error='m' | GSL math error. |
 -test: the ANOVA approach, which is typically built around the claim that all effects but the mean are zero; and the more general regression form, which allows for any set of linear claims about the data. If you send a
-test: the ANOVA approach, which is typically built around the claim that all effects but the mean are zero; and the more general regression form, which allows for any set of linear claims about the data. If you send a NULL contrast set, I will generate the set of linear contrasts that are equivalent to the ANOVA-type approach. This is why the top row of the default  matrix is missing: there is no hypothesis test about the coefficient for the constant term. See the example below.
matrix is missing: there is no hypothesis test about the coefficient for the constant term. See the example below. Runs an F-test specified by q and c. See the chapter on hypothesis testing in Modeling With Data, p 309, which will tell you that:
![\[{N-K\over q} {({\bf Q}'\hat\beta - {\bf c})' [{\bf Q}' ({\bf X}'{\bf X})^{-1} {\bf Q}]^{-1} ({\bf Q}' \hat\beta - {\bf c}) \over {\bf u}' {\bf u} } \sim F_{q,N-K},\]](form_93.png)
and that's what this function is based on.
| est | An apop_model that you have already calculated. (No default) |
| contrast | An apop_data set whose matrix represents  and whose vector represents and whose vector represents  . Each row represents a hypothesis. (Defaults: if matrix is . Each row represents a hypothesis. (Defaults: if matrix is NULL, it is set to the identity matrix with the top row missing. If the vector is NULL, it is set to a zero matrix of length equal to the height of the contrast matrix. Thus, if the entire apop_data set is NULL or omitted, we are testing the hypothesis that all but 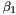 are zero.) are zero.) |
apop_data set with a few variants on the confidence with which we can reject the joint hypothesis. | out->error='a' | Allocation error. |
| out->error='d' | dimension-matching error. |
| out->error='i' | matrix inversion error. |
| out->error='m' | GSL math error. |
 -test: the ANOVA approach, which is typically built around the claim that all effects but the mean are zero; and the more general regression form, which allows for any set of linear claims about the data. If you send a
-test: the ANOVA approach, which is typically built around the claim that all effects but the mean are zero; and the more general regression form, which allows for any set of linear claims about the data. If you send a NULL contrast set, I will generate the set of linear contrasts that are equivalent to the ANOVA-type approach. This is why the top row of the default  matrix is missing: there is no hypothesis test about the coefficient for the constant term. See the example below.
matrix is missing: there is no hypothesis test about the coefficient for the constant term. See the example below. | long double apop_generalized_harmonic | ( | int | N, |
| double | s | ||
| ) |
Calculate 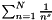
N is zero or negative, return NaN. Notify the user if apop_opts.verbosity >=0For example:
| apop_data * apop_histograms_test_goodness_of_fit | ( | apop_model * | observed, |
| apop_model * | expected | ||
| ) |
Test the goodness-of-fit between two apop_pmf models.
Let 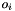 be the
be the 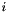 th observed bin and
th observed bin and  the expected value of that bin; then under typical assumptions, $
the expected value of that bin; then under typical assumptions, $  .
.
If you send two histograms, I assume that the histograms are synced: for PMFs, you've used apop_data_to_bins to generate two histograms using the same binspec, or you've used apop_data_pmf_compress to guarantee that each observation value appears exactly once in each data set.
In any case, all values in the observed set must appear in the expected set with nonzero weight; otherwise this will return a  statistic of
statistic of GSL_POSINF, indicating that it is impossible for the observed data to have been drawn from the expected distribution.
apop_opts.verbose >=1 I will show a warning. | apop_data * apop_jackknife_cov | ( | apop_data * | in, |
| apop_model * | model | ||
| ) |
Give me a data set and a model, and I'll give you the jackknifed covariance matrix of the model parameters.
The basic algorithm for the jackknife (glossing over the details): create a sequence of data sets, each with exactly one observation removed, and then produce a new set of parameter estimates using that slightly shortened data set. Then, find the covariance matrix of the derived parameters.
| in | The data set. An apop_data set where each row is a single data point. |
| model | An apop_model, that will be used internally by apop_estimate. |
| out->error=='n' | NULL input data. |
apop_data set whose matrix element is the estimated covariance matrix of the parameters. For example:
| long double apop_kl_divergence | ( | apop_model * | from, |
| apop_model * | to, | ||
| int | draw_ct, | ||
| gsl_rng * | rng | ||
| ) |
Kullback-Leibler divergence.
This measure of the divergence of one distribution from another has the form 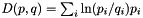 . Notice that it is not a distance, because there is an asymmetry between
. Notice that it is not a distance, because there is an asymmetry between  and
and  , so one can expect that
, so one can expect that  .
.
| from | the  in the above formula. (No default; must not be in the above formula. (No default; must not be NULL) |
| to | the 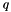 in the above formula. (No default; must not be in the above formula. (No default; must not be NULL) |
| draw_ct | If I do the calculation via random draws, how many? (Default = 1e5) |
| rng | A gsl_rng. If NULL or number of threads is greater than 1, I'll take care of the RNG; see apop_rng_get_thread. (Default = NULL) |
This function can take empirical histogram-type models (apop_pmf) or continuous models like apop_loess or apop_normal.
If the from distribution is a PMF (determined by checking whether its p function is that of apop_pmf), then I'll step through it for the points in the summation.
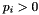 but
but 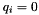 , then the function returns
, then the function returns GSL_NEGINF. If apop_opts.verbose >=1 I print a message as well.If the from distribution is not a PMF, then I will take draw_ct random draws from from and evaluate at those points.
apop_opts.verbose = 3 for observation-by-observation info.| long double apop_linear_constraint | ( | gsl_vector * | beta, |
| apop_data * | constraint, | ||
| double | margin | ||
| ) |
This is designed to be called from within the constraint method of your apop_model. Just write the constraint vector+matrix and this will do the rest. See Setting Constraints for detailed discussion.
| beta | The proposed vector about to be tested. No default, must not be NULL. |
| constraint | A vector/matrix pair [v | m1 m2 ... mn] where each row is interpreted as a less-than inequality:  . For example, say your constraints are . For example, say your constraints are 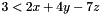 and and  is positive, i.e. is positive, i.e. 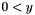 . Allocate and fill the matrix representing these two constraints via: . Default: each elements is greater than zero. For three parameters this would be equivalent to setting . Allocate and fill the matrix representing these two constraints via: . Default: each elements is greater than zero. For three parameters this would be equivalent to setting |
| margin | If zero, then this is a >= constraint, otherwise I will return a point this amount within the borders. You could try GSL_DBL_EPSILON, which is the smallest value a double can hold, or something like 1e-3. Default = 0. |
beta is shifted by margin (Euclidean distance) to meet the constraints.| double apop_log_likelihood | ( | apop_data * | d, |
| apop_model * | m | ||
| ) |
Find the log likelihood of a data/parametrized model pair.
| d | The data |
| m | The parametrized model, which must have either a log_likelihood or a p method. |
| apop_data * apop_map | ( | apop_data * | in, |
| apop_fn_d * | fn_d, | ||
| apop_fn_v * | fn_v, | ||
| apop_fn_r * | fn_r, | ||
| apop_fn_dp * | fn_dp, | ||
| apop_fn_vp * | fn_vp, | ||
| apop_fn_rp * | fn_rp, | ||
| apop_fn_dpi * | fn_dpi, | ||
| apop_fn_vpi * | fn_vpi, | ||
| apop_fn_rpi * | fn_rpi, | ||
| apop_fn_di * | fn_di, | ||
| apop_fn_vi * | fn_vi, | ||
| apop_fn_ri * | fn_ri, | ||
| void * | param, | ||
| int | inplace, | ||
| char | part, | ||
| int | all_pages | ||
| ) |
Apply a function to every element of a data set, matrix or vector; or, apply a vector-taking function to every row or column of a matrix.
Your function could take any combination of a gsl_vector, a double, an apop_data, a parameter set, and the position of the element in the vector or matrix. As such, the function takes twelve function inputs, one for each combination of vector/matrix, params/no params, index/no index. Fortunately, because this function uses the Designated initializers syntax for inputs, you will specify only one.
For example, here is a function that will cut off each element of the input data to between 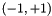 . It takes in a lone
. It takes in a lone double and a parameter in a void*, so it gets sent to apop_map via .fn_dp=cutoff.
| fn_v | A function of the form double your_fn(gsl_vector *in) |
| fn_d | A function of the form double your_fn(double in) |
| fn_r | A function of the form double your_fn(apop_data *in) |
| fn_vp | A function of the form double your_fn(gsl_vector *in, void *param) |
| fn_dp | A function of the form double your_fn(double in, void *param) |
| fn_rp | A function of the form double your_fn(apop_data *in, void *param) |
| fn_vpi | A function of the form double your_fn(gsl_vector *in, void *param, int index) |
| fn_dpi | A function of the form double your_fn(double in, void *param, int index) |
| fn_rpi | A function of the form double your_fn(apop_data *in, void *param, int index) |
| fn_vi | A function of the form double your_fn(gsl_vector *in, int index) |
| fn_di | A function of the form double your_fn(double in, int index) |
| fn_ri | A function of the form double your_fn(apop_data *in, int index) |
| in | The input data set. If NULL, I'll return NULL immediately. |
| param | A pointer to the parameters to be passed to those function forms taking a *param. |
| part | Which part of the apop_data struct should I use?'v'==Just the vector 'm'==Every element of the matrix, in turn 'a'==Both 'v' and 'm' 'r'==Apply a function gsl_vector  double to each row of the matrix'c'==Apply a function gsl_vector  double to each column of the matrixDefault is 'a', but notice that I'll ignore a NULL vector or matrix, so if your data set has only a vector or only a matrix, that's what I'll use. |
| all_pages | If 'y', then follow the more pointer to subsequent pages. If 'n', handle only the first page of data. Default: 'n'. |
| inplace | If 'n' (the default), generate a new apop_data set for output, which will contain the mapped values (and the names from the original set). If 'y', modify in place. The double  double versions, 'v', 'm', and 'a', write to exactly the same location as before. The gsl_vector  double versions, 'r', and 'c', will write to the vector. Be careful: if you are writing in place and there is already a vector there, then the original vector is lost.If 'v' (as in void), return NULL. (Default = 'n') |
| out->error='p' | missing or mismatched parts error, such as NULL matrix when you sent a function acting on the matrix element. |
r in them, like fn_ri, are row-by-row. I'll use Apop_r to get each row in turn, and send it to the function. The first implication is that your function should be expecting a apop_data set with exactly one row in it. The second is that part is ignored: it only makes sense to go row-by-row. r functions, if you set inplace='y', then you will be modifying your input data set, row by row; if you set inplace='n', then I will return an apop_data set whose vector element is as long as your data set (i.e., as long as the longest of your text, vector, or matrix parts). omp_set_num_threads(n) using 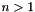 , split the data set into as many chunks as you specify and process them simultaneously. You need to watch out for the usual hang-ups about multithreaded programming, but if your data is iid, and each row's processing is independent of the others, you should have no problems. Bear in mind that generating threads takes some small overhead, so simple cases like adding a few hundred numbers will actually be slower when threading.
, split the data set into as many chunks as you specify and process them simultaneously. You need to watch out for the usual hang-ups about multithreaded programming, but if your data is iid, and each row's processing is independent of the others, you should have no problems. Bear in mind that generating threads takes some small overhead, so simple cases like adding a few hundred numbers will actually be slower when threading. | double apop_map_sum | ( | apop_data * | in, |
| apop_fn_d * | fn_d, | ||
| apop_fn_v * | fn_v, | ||
| apop_fn_r * | fn_r, | ||
| apop_fn_dp * | fn_dp, | ||
| apop_fn_vp * | fn_vp, | ||
| apop_fn_rp * | fn_rp, | ||
| apop_fn_dpi * | fn_dpi, | ||
| apop_fn_vpi * | fn_vpi, | ||
| apop_fn_rpi * | fn_rpi, | ||
| apop_fn_di * | fn_di, | ||
| apop_fn_vi * | fn_vi, | ||
| apop_fn_ri * | fn_ri, | ||
| void * | param, | ||
| char | part, | ||
| int | all_pages | ||
| ) |
A function that effectively calls apop_map and returns the sum of the resulting elements. Thus, this function returns a double. See the apop_map page for details of the inputs, which are the same here, except that inplace doesn't make sense—this function will always just add up the input function outputs.
NULL. If apop_opts.verbose >= 2 print a warning. | void apop_matrix_apply | ( | gsl_matrix * | m, |
| void(*)(gsl_vector *) | fn | ||
| ) |
Apply a function to every row of a matrix. The function that you input takes in a gsl_vector and returns nothing. apop_matrix_apply will produce a vector view of each row, and send each row to your function.
| m | The matrix |
| fn | A function of the form void fn(gsl_vector* in) which may modify the data at the in pointer in place. |
NULL, this is a no-op and returns immediately. | void apop_matrix_apply_all | ( | gsl_matrix * | in, |
| void(*)(double *) | fn | ||
| ) |
Applies a function to every element in a matrix (as opposed to every row)
| in | The matrix whose elements will be inputs to the function |
| fn | A function with a form like void f(double *in) which may modify the data at the in pointer in place. |
NULL, this is a no-op and returns immediately. | gsl_matrix * apop_matrix_copy | ( | const gsl_matrix * | in | ) |
Copy one gsl_matrix to another. That is, all data are duplicated. Unlike gsl_matrix_memcpy, this function allocates and returns the destination, so you can use it like this:
| in | the input data |
gsl_matrix_alloc fails, returns NULL. | double apop_matrix_determinant | ( | const gsl_matrix * | in | ) |
Find the determinant of a matrix. The in matrix is not destroyed in the process.
See also apop_matrix_inverse , or apop_det_and_inv to do both at once.
| in | The matrix to be determined. |
| gsl_matrix * apop_matrix_inverse | ( | const gsl_matrix * | in | ) |
Inverts a matrix. The in matrix is not destroyed in the process. You may want to call apop_matrix_determinant first to check that your input is invertible, or use apop_det_and_inv to do both at once.
| in | The matrix to be inverted. |
| int apop_matrix_is_positive_semidefinite | ( | gsl_matrix * | m, |
| char | semi | ||
| ) |
Test whether the input matrix is positive semidefinite (PSD).
A covariance matrix will always be PSD, so this function can tell you whether your matrix is a valid covariance matrix.
Consider the 1x1 matrix in the upper left of the input, then the 2x2 matrix in the upper left, on up to the full matrix. If the matrix is PSD, then each of these has a positive determinant. This function thus calculates 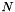 determinants for an
determinants for an 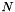 x
x 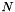 matrix.
matrix.
| m | The matrix to test. If NULL, I will return zero—not PSD. |
| semi | If anything but 's', check for positive definite, not semidefinite. (default 's') |
See also apop_matrix_to_positive_semidefinite, which will change the input to something PSD.
| gsl_vector * apop_matrix_map | ( | const gsl_matrix * | m, |
| double(*)(gsl_vector *) | fn | ||
| ) |
Map a function onto every row of a matrix. The function that you input takes in a gsl_vector and returns a double. This function will produce a sequence of vector views of each row of the input matrix, and send each to your function. It will output a gsl_vector holding your function's output for each row.
| m | The matrix |
| fn | A function of the form double fn(gsl_vector* in) |
gsl_vector with the corresponding value for each row.NULL matrix, I return NULL. | gsl_matrix * apop_matrix_map_all | ( | const gsl_matrix * | in, |
| double(*)(double) | fn | ||
| ) |
Maps a function to every element in a matrix (as opposed to every row).
| in | The matrix whose elements will be inputs to the function |
| fn | A function with a form like double f(double in). |
NULL matrix, I return NULL. | double apop_matrix_map_all_sum | ( | const gsl_matrix * | in, |
| double(*)(double) | fn | ||
| ) |
Like apop_matrix_map_all, but returns the sum of the resulting mapped function. For example, apop_matrix_map_all_sum(v, isnan) returns the number of elements of m that are NaN.
NULL matrix, I return the sum of zero items: zero. | double apop_matrix_map_sum | ( | const gsl_matrix * | in, |
| double(*)(gsl_vector *) | fn | ||
| ) |
Like apop_matrix_map, but returns the sum of the resulting mapped vector. For example, let log_like be a function that returns the log likelihood of an input vector; then apop_matrix_map_sum(m, log_like) returns the total log likelihood of the rows of m.
NULL matrix, I return the sum of zero items: zero. | double apop_matrix_mean | ( | const gsl_matrix * | data | ) |
Returns the mean of all elements of a matrix.
| data | The matrix to be averaged. If NULL, return zero. |
| void apop_matrix_mean_and_var | ( | const gsl_matrix * | data, |
| double * | mean, | ||
| double * | var | ||
| ) |
Returns the mean and population variance of all elements of a matrix.
NULL, return 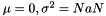 .
. 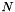 ). If you want sample variance, multiply the result by
). If you want sample variance, multiply the result by 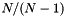 :
: | data | the matrix to be averaged. |
| mean | where to put the mean to be calculated. |
| var | where to put the variance to be calculated. |
| apop_data * apop_matrix_pca | ( | gsl_matrix * | data, |
| int const | dimensions_we_want | ||
| ) |
Principal component analysis: hand in a matrix and (optionally) a number of desired dimensions, and I'll return a data set where each column of the matrix is an eigenvector. The columns are sorted, so column zero has the greatest weight. The vector element of the data set gives the weights.
You may also specify the number of elements your principal component space should have. If this is equal to the rank of the space in which the input data lives, then the sum of weights will be one. If the dimensions desired is less than that (probably so you can prepare a plot), then the weights will be accordingly smaller, giving you an indication of how much variation these dimensions explain.
| data | The input matrix. I modify int in place so that each column has mean zero. (No default. If NULL, return NULL and print a warning iff apop_opts.verbose >= 1.) |
| dimensions_we_want | The singular value decomposition will return this many of the eigenvectors with the largest eigenvalues. (default: the size of the covariance matrix, i.e. data->size2) |
The data set's vector will be the largest eigenvalues, scaled by the total of all eigenvalues (including those that were thrown out). The sum of these returned values will give you the percentage of variance explained by the factor analysis.
| out->error=='a' | Allocation error. |
| void apop_matrix_print | ( | const gsl_matrix * | data, |
| Output_declares | |||
| ) |
Print a gsl_matrix to the screen, a file, a pipe, or a database table.
| gsl_matrix * apop_matrix_realloc | ( | gsl_matrix * | m, |
| size_t | newheight, | ||
| size_t | newwidth | ||
| ) |
This function will resize a gsl_matrix to a new height or width.
Data in the matrix will be retained. If the new height or width is smaller than the old, then data in the later rows/columns will be cropped away (in a non–memory-leaking manner). If the new height or width is larger than the old, then new cells will be filled with garbage; it is your responsibility to zero out or otherwise fill new rows/columns before use.
reallocs can take a noticeable amount of time. You are encouraged to determine the size of your data beforehand and avoid writing for loops that reallocate the matrix at every iteration. gsl_matrix is a versatile struct that can represent submatrices and other cuts from parent data. Resizing a subset of a parent matrix makes no sense, so return NULL and print a warning if asked to resize a view of a matrix.| m | The already-allocated matrix to resize. If you give me NULL, this becomes equivalent to gsl_matrix_alloc |
| newheight,newwidth | The height and width you'd like the matrix to be. |
| gsl_matrix * apop_matrix_stack | ( | gsl_matrix * | m1, |
| gsl_matrix const * | m2, | ||
| char | posn, | ||
| char | inplace | ||
| ) |
Put the first matrix either on top of or to the right of the second matrix. Returns a new matrix, meaning that at the end of this function, until you gsl_matrix_free() the original matrices, you will be taking up twice as much memory. Plan accordingly.
| m1 | the upper/rightmost matrix (default: NULL, in which case this copies m2) |
| m2 | the second matrix (default: NULL, in which case m1 is returned) |
| posn | If 'r', stack rows on top of other rows. If 'c' stack columns next to columns. (default: 'r') |
| inplace | If 'y', use apop_matrix_realloc to modify m1 in place; see the caveats on that function. Otherwise, allocate a new matrix, leaving m1 undisturbed. (default: 'n') |
m1.For example, here is a function to merge four matrices into a single two-part-by-two-part matrix. The original matrices are unchanged.
| long double apop_matrix_sum | ( | const gsl_matrix * | m | ) |
Returns the sum of the elements of a matrix. Occasionally convenient.
| m | the matrix to be summed. |
| double apop_matrix_to_positive_semidefinite | ( | gsl_matrix * | m | ) |
This function takes in a matrix and converts it in place to the `closest' positive semidefinite matrix.
| m | On input, any matrix; on output, a positive semidefinite matrix. If NULL, return NaN and print an error. |
| void apop_maximum_likelihood | ( | apop_data * | data, |
| apop_model * | dist | ||
| ) |
Find the likelihood-maximizing parameters of a model given data.
estimate method.| data | An apop_data set. |
| dist | The apop_model object: apop_gamma, apop_probit, apop_zipf, &c. You can add an apop_mle_settings struct to it (Apop_model_add_group(your_model, apop_mle, .verbose=1, .method="PR cg", and_so_on)). |
->info element of the post-estimation struct. Get elements via, e.g.: | apop_model * apop_ml_impute | ( | apop_data * | d, |
| apop_model * | mvn | ||
| ) |
Impute the most likely data points to replace NaNs in the data, and insert them into the given data. That is, the data set is modified in place.
How it works: this uses the machinery for apop_model_fix_params. The only difference is that this searches over the data space and takes the parameter space as fixed, while basic fix params model searches parameters and takes data as fixed. So this function just does the necessary data-parameter switching to make that happen.
| d | The data set. It comes in with NaNs and leaves entirely filled in. |
| mvn | A parametrized apop_model from which you expect the data was derived. if NULL, then I'll use the Multivariate Normal that best fits the data after listwise deletion. |
| apop_model * apop_model_clear | ( | apop_data * | data, |
| apop_model * | model | ||
| ) |
Set up the parameters and info elements of the apop_model:
At close, the input model has parameters of the correct size.
| data | If your params vary with the size of the data set, then the function needs a data set to calibrate against. Otherwise, it's OK to set this to NULL. |
| model | The model whose output elements will be modified. |
| outmodel->error=='d' | dimension error. |
| apop_model * apop_model_copy | ( | apop_model * | in | ) |
Outputs a copy of the apop_model input.
| in | The model to be copied |
parameters (if not NULL, copied via apop_data_copy).in.more_size > 0 I memcpy the more pointer from the original data set. in->data is not copied, but is also pointed to.| out->error=='a' | Allocation error. In extreme cases, where there aren't even a few hundred bytes available, I will return NULL. |
| out->error=='s' | Error copying settings groups. |
| out->error=='p' | Error copying parameters or info page; the given apop_data struct may be NULL or may have its own ->error element. |
| apop_data * apop_model_draws | ( | apop_model * | model, |
| int | count, | ||
| apop_data * | draws | ||
| ) |
Make a set of random draws from a model and write them to an apop_data set.
| model | The model from which draws will be made. Must already be prepared and/or estimated. |
| count | The number of draws to make. If draw_matrix is not NULL, then this is ignored and count=draw_matrix->matrix->size1. default=1000. |
| draws | If not NULL, a pre-allocated data set whose matrix element will be filled with draws. |
size draws. If draw_matrix!=NULL, then return a pointer to it.| out->error=='m' | Input model isn't good for making draws: it is NULL, or m->dsize=0. |
| out->error=='s' | You gave me a draws matrix, but its size is less than the size of a single draw from the data, model->dsize. |
| out->error=='d' | Trouble drawing from the distribution for at least one row. That row is set to all NAN. |
NULL apop_data set, but its matrix element is NULL, when apop_opts.verbose>=1. Here is a two-line program to draw a different set of ten Standard Normals on every run (provided runs are more than a second apart):
| long double apop_model_entropy | ( | apop_model * | in, |
| int | draws | ||
| ) |
Calculate the entropy of a model:  , which is the expected value of
, which is the expected value of  .
.
The default method is to make draws using apop_model_draws, then evaluate the log likelihood at those points using the model's log_likelihood method.
There are a number of routines for specific models, inlcuding the apop_normal and apop_pmf models.
 :
: apop_model_entropy(my_model)/log(2).| in | A parameterized apop_model. That is, you have already used apop_estimate or apop_model_set_parameters to estimate/set the model parameters. |
| draws | If using the default method of making random draws, how many random draws to make (default=1,000) |
Sample code:
| apop_model * apop_model_fix_params | ( | apop_model * | model_in | ) |
Produce a model based on another model, but with some of the parameters fixed at a given value.
You will send me the model whose parameters you want fixed, with the parameters element set as follows. For the fixed parameters, simply give the values to which they will be fixed. Set the free parameters to NaN.
For example, here is a Binomial distribution with a fixed  but
but  allowed to float freely:
allowed to float freely:
The output is an apop_model that can be estimated, Bayesian updated, et cetera.
estimate method always uses an MLE, and it never calls the base model's estimate method. estimate method. Otherwise, I'll set my own. more pointer of the parameters for additional pages and NaNs on those pages.Here is a sample program. It produces a few thousand draws from a Multivariate Normal distribution, and then tries to recover the means given a var/covar matrix fixed at the correct variance.
| model_in | The base model |
| apop_model * apop_model_fix_params_get_base | ( | apop_model * | fixed_model | ) |
The apop_model_fix_params function produces a model that has only the non-fixed parameters of the model. After estimation of the fixed-parameter model, this function fills the parameters element of the base model and returns a pointer to the base model.
| void apop_model_free | ( | apop_model * | free_me | ) |
Free an apop_model structure.
parameters and settings are freed. These are the elements that are copied by apop_model_copy. data element is not freed, because the odds are you still need it. free_me->more_size is positive, the function runs free(free_me->more). But it has no idea what the more element contains; if it points to other structures (like an apop_data set), you need to free them before calling this function. free_me is NULL, this does nothing.| free_me | A pointer to the model to be freed. |
| apop_data * apop_model_hessian | ( | apop_data * | data, |
| apop_model * | model, | ||
| double | delta | ||
| ) |
Numerically estimate the matrix of second derivatives of the parameter values, via a series of re-evaluations at small differential steps. [Therefore, it may be expensive to do this for a very computationally-intensive model.]
| data | The apop_data at which the model was estimated (default: NULL) |
| model | The apop_model, with parameters already estimated (no default, must not be NULL) |
| delta | the step size for the differentials. (default: 1e-3, but see below) |
delta element. If that is also missing, use the default of 1e-3. | apop_model * apop_model_metropolis | ( | apop_data * | d, |
| gsl_rng * | rng, | ||
| apop_model * | m | ||
| ) |
Use Metropolis-Hastings Markov chain Monte Carlo to make draws from the given model.
The basic storyline is that draws are made from a proposal distribution, and the likelihood of your model given your data and the drawn parameters evaluated. At each step, a new set of proposal parameters are drawn, and if they are more likely than the previous set the new proposal is accepted as the next step, else with probability (prob of new params)/(prob of old params), they are accepted as the next step anyway. Otherwise the last accepted proposal is repeated.
The output is an apop_pmf model with a data set listing the draws that were accepted, including those repetitions. The output model is modified so that subsequent draws are one more step from the Markov chain, via apop_model_metropolis_draw.
| d | The apop_data set used for evaluating the likelihood of a proposed parameter set. |
| rng | A gsl_rng, probably allocated via apop_rng_alloc. (Default: an RNG from apop_rng_get_thread) |
| m | The apop_model from which parameters are being drawn. (No default; must not be NULL) |
draw method that returns another step from the Markov chain with each draw.| out->error='c' | Proposal was outside of a constraint; see below. |
constraint element of the model you input, then the proposal is thrown out and a new one selected. By the default proposal distribution, this is not mathematically correct (it breaks detailed balance), and values near the constraint will be oversampled. The output model will have outmodel->error=='c'. It is up to you to decide whether the resulting distribution is good enough for your purposes or whether to take the time to write a custom proposal and step function to accommodate the constraint.Attach an apop_mcmc_settings group to your model to specify the proposal distribution, burnin, and other details of the search. See the apop_mcmc_settings documentation for details.
base_adapt_fn in the apop_mcmc_settings group to a do-nothing function, or one that damps its adaptation as  .
. gibbs_chunks element of the apop_mcmc_settings group. If you set gibbs_chunks='a', all parameters are drawn as a set, and accepted/rejected as a set. The variances are adapted at an identical rate. If you set gibbs_chunks='i', then each scalar parameter is assigned its own proposal distribution, which is adapted at its own pace. With gibbs_chunks='b' (the default), then each of the vector, matrix, and weights of your model's parameters are drawn/accepted/adapted as a block (and so on to additional chunks if your model has ->more pages). This works well for complex models which naturally break down into subsets of parameters. NULL parameters, I will allocate them. That means you can use one of the stock models that ship with Apophenia. If I need to run the model's prep routine to get the size of the parameters, then I will make a copy of the likelihood model, run prep, and then allocate parameters for that copy of a model. parameters element of your likelihood model has the last accepted parameter proposal. apop_opts.verbose=2 or greater, I will report the accept rate of the M-H sampler. It is a common rule of thumb to select a proposal so that this is between 20% and 50%. Set apop_opts.verbose=3 to see the stream of proposal points, their likelihoods, and the acceptance odds. You may want to set apop_opts.log_file=fopen("yourlog", "w") first.| int apop_model_metropolis_draw | ( | double * | out, |
| gsl_rng * | rng, | ||
| apop_model * | model | ||
| ) |
The draw method for models estimated via apop_model_metropolis.
That method produces an apop_pmf, typically with a few thousand draws from the model in a batch. If you want to get a single next step from the Markov chain, use this.
A Markov chain works by making a new draw and then accepting or rejecting the draw. If the draw is rejected, the last value is reported as the next step in the chain. Users sometimes mitigate this repetition by making a batch of draws (say, ten at a time) and using only the last.
If you run this without first running apop_model_metropolis, I will run it for you, meaning that there will be an initial burn-in period before the first draw that can be reported to you. That run is done using model->data as input.
| out | An array of doubles, which will hold the draw, in the style of apop_draw. |
| rng | A gsl_rng, already initialized, probably via apop_rng_alloc. |
| model | A model which was probably already run through apop_model_metropolis. |
out is filled with the next step in the Markov chain. The ->data element of the PMF model is extended to include the additional steps in the chain. If a proposal failed the model constraints, then return 1; else return 0. See the notes in the documentation for apop_model_metropolis.base_model in the mcmc settings group == the parent model. | apop_data * apop_model_numerical_covariance | ( | apop_data * | data, |
| apop_model * | model, | ||
| double | delta | ||
| ) |
Produce the covariance matrix for the parameters of an estimated model via the derivative of the score function at the parameter. I.e., I find the second derivative via apop_model_hessian , and take the negation of the inverse.
I follow Efron and Hinkley in using the estimated information matrix—the value of the information matrix at the estimated value of the score—not the expected information matrix that is the integral over all possible data. See Pawitan 2001 (who cribbed a little off of Efron and Hinkley) or Klemens 2008 (who directly cribbed off of both) for further details.
| data | The data by which your model was estimated |
| model | A model whose parameters have been estimated. |
| delta | The differential by which to step for sampling changes. (default: 1e-3, but see below) |
"<Covariance>" page, I'll set it to the result as well [i.e., I won't overwrite an existing covariance page].delta element. If that is also missing, use the default of 1e-3. | void apop_model_print | ( | apop_model * | model, |
| FILE * | output_pipe | ||
| ) |
Print the results of an estimation for a human to look over.
| model | The model whose information should be displayed (No default. If NULL, print NULL) |
| output_pipe | The output stream. Default: stdout. If you'd like something else, use fopen. E.g.: |
When building a special print method, all output should fprintf to the input FILE* handle. Apophenia's output routines also accept a file handle; e.g., if the file handle is named out, then if the thismodel print method uses apop_data_print to print the parameters, it must do so via a form like apop_data_print(thismodel->parameters, .output_pipe=ap).
Your print method can use both by masking itself for a few lines:
| apop_model * apop_model_to_pmf | ( | apop_model * | model, |
| apop_data * | binspec, | ||
| long int | draws, | ||
| int | bin_count | ||
| ) |
Make random draws from an apop_model, and bin them using a binspec in the style of apop_data_to_bins. If you have a data set that used the same binspec, you now have synced histograms, which you can plot or sensibly test hypotheses about.
| binspec | A description of the bins in which to place the draws; see apop_data_to_bins. (default: as in apop_data_to_bins.) |
| model | The model to be drawn from. Because this function works via random draws, the model needs to have a draw method. (No default) |
| draws | The number of random draws to make. (arbitrary default = 10,000) |
| bin_count | If no bin spec, the number of bins to use (default: as per apop_data_to_bins,  ) ) |
apop_data_free(output_model->data) to prevent memory leaks). The weights on the data set are normalized to sum to one.| long double apop_multivariate_gamma | ( | double | a, |
| int | p | ||
| ) |
The multivariate generalization of the Gamma distribution.
![\[ \Gamma_p(a)= \pi^{p(p-1)/4}\prod_{j=1}^p \Gamma\left[ a+(1-j)/2\right]. \]](form_88.png)
Because  is undefined for
is undefined for  , this function returns
, this function returns NAN when  takes on one of those values.
takes on one of those values.
See also apop_multivariate_lngamma, which is more numerically stable in most cases.
| long double apop_multivariate_lngamma | ( | double | a, |
| int | p | ||
| ) |
The log of the multivariate generalization of the Gamma; see also apop_multivariate_gamma.
| int apop_name_add | ( | apop_name * | n, |
| char const * | add_me, | ||
| char | type | ||
| ) |
Adds a name to the apop_name structure. Puts it at the end of the given list.
| n | An existing, allocated apop_name structure. |
| add_me | A string. If NULL, do nothing; return -1. |
| type | 'r': add a row name 'c': add a matrix column name 't': add a text column name 'h': add a title (i.e., a header). 'v': add (or overwrite) the vector name |
add_me is NULL, return -1. | apop_name * apop_name_alloc | ( | void | ) |
Allocates a name structure
malloc fails, return NULL.Because apop_data_alloc uses this to set up its output, you will rarely if ever need to call this function explicitly. You may want to use it if wrapping a gsl_matrix into an apop_data set. For example, to put a title on a vector:
Copy one apop_name structure to another. That is, all data is duplicated.
Used internally by apop_data_copy, but sometimes useful by itself. For example, say that we have an apop_data struct named d and a gsl_matrix of the same dimensions named m; we could give m the labels from d for printing:
| in | The input names |
| int apop_name_find | ( | const apop_name * | n, |
| const char * | name, | ||
| const char | type | ||
| ) |
Finds the position of an element in a list of names.
The function uses POSIX's strcasecmp, and so does case-insensitive search the way that function does.
| n | the apop_name object to search. |
| name | the name you seek; see above. |
| type | 'c' (=column), 'r' (=row), or 't' (=text). Default is 'c'. |
findme. If 'c', then this may be -1, meaning the vector name. If not found, returns -2. On error, e.g. name==NULL, returns -2. | void apop_name_print | ( | apop_name * | n | ) |
Prints the given list of names to stdout. Useful for debugging.
| n | The apop_name structure |
Append one list of names to another.
If the first list is empty, then this is a copy function.
| n1 | The first set of names (no default, must not be NULL) |
| nadd | The second set of names, which will be appended after the first. (no default. If NULL, a no-op.) |
| type1 | Either 'c', 'r', 't', or 'v' stating whether you are merging the columns, rows, text, or vector. If 'v', then ignore typeadd and just overwrite the target vector name with the source name. (default: 'r') |
| typeadd | Either 'c', 'r', 't', or 'v' stating whether you are merging the columns, rows, or text. If 'v', then overwrite the target with the source vector name. (default: type1) |
| gsl_vector * apop_numerical_gradient | ( | apop_data * | data, |
| apop_model * | model, | ||
| double | delta | ||
| ) |
A wrapper around the GSL's one-dimensional gsl_deriv_central to find a numeric differential for each dimension of the input apop_model's log likelihood (or p if log_likelihood is NULL).
| data | The apop_data set to use for all evaluations. |
| model | The apop_model, expressing the function whose derivative is sought. The gradient is taken via small changes along the model parameters. |
| delta | The size of the differential. (default: 1e-3, but see below) |
delta element. If that is also missing, use the default of 1e-3. | double apop_p | ( | apop_data * | d, |
| apop_model * | m | ||
| ) |
Find the probability of a data/parametrized model pair.
| d | The data |
| m | The parametrized model, which must have either a log_likelihood or a p method. |
| apop_data * apop_paired_t_test | ( | gsl_vector * | a, |
| gsl_vector * | b | ||
| ) |
Answers the question: with what confidence can I say that the mean difference between the two columns is zero?
If apop_opts.verbose >=2, then display some information, like the mean/var/count for both vectors and the t statistic, to stderr.
| a | A column of data |
| b | A matched column of data |
mean left - right: the difference in means; if positive, first vector has larger mean, and one-tailed test is testing 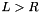 , else reverse if negative.
, else reverse if negative.t statistic: used for the testdf: degrees of freedomp value, 1 tail: the p-value for a one-tailed test that one vector mean is greater than the other.confidence, 1 tail: 1- p value.p value, 2 tail: the p-value for the two-tailed test that left mean = right mean.confidence, 2 tail: 1-p value| apop_model * apop_parameter_model | ( | apop_data * | d, |
| apop_model * | m | ||
| ) |
Get a model describing the distribution of the given parameter estimates.
For many models, the parameter estimates are well-known, such as the 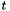 -distribution of the parameters for OLS.
-distribution of the parameters for OLS.
For models where the distribution of  is not known, if you give me data, I will return an apop_normal or apop_multivariate_normal model, using the parameter estimates as mean and apop_bootstrap_cov for the variances.
is not known, if you give me data, I will return an apop_normal or apop_multivariate_normal model, using the parameter estimates as mean and apop_bootstrap_cov for the variances.
If you don't give me data, then I will assume that this is a stochastic model where re-running the model will produce different parameter estimates each time. In this case, I will run the model 1e4 times and return a apop_pmf model with the resulting parameter distributions.
Before calling this, I expect that you have already run apop_estimate to produce  .
.
The apop_pm_settings structure dictates details of how the model is generated. For example, if you want only the distribution of the third parameter, and you know the distribution will be a PMF generated via random draws, then set settings and call the model via:
Some useful parts of apop_pm_settings:
index gives the position of the parameter (in apop_data_pack order) in which you are interested. Thus, if this is zero or more, then you will get a univariate output distribution describing a single parameter. If index == -1, then I will give you the multivariate distribution across all parameters. The default is zero (i.e. the univariate distribution of the zeroth parameter). draws If there is no closed-form solution and bootstrap is inappropriate, then the last resort is a large numbr of random draws of the model, summarized into a PMF. Default: 1,000 draws. rng If the method requires random draws, then use this. If you provide NULL and one is needed, I provide one for you via apop_rng_get_thread.The default is via resampling as above, but special-case calculations for certain models are held in a vtable; see Registering new methods in vtables for details. The typedef new functions must conform to and the hash used for lookups are:
| apop_data * apop_predict | ( | apop_data * | d, |
| apop_model * | m | ||
| ) |
A prediction supplies E(a missing value | original data, already-estimated parameters, and other supplied data elements ).
For a regression, one would first estimate the parameters of the model, then supply a row of predictors X. The value of the dependent variable  is unknown, so the system would predict that value.
is unknown, so the system would predict that value.
For a univariate model (i.e. a model in one-dimensional data space), there is only one variable to omit and fill in, so the prediction problem reduces to the expected value: E(a missing value | original data, already-estimated parameters). [In some models, this may not be the expected value, but is a best value for the missing item using some other meaning of `best'.]
In other cases, prediction is the missing data problem: for three-dimensional data, you may supply the input (34, NaN, 12), and the parameterized model provides the most likely value of the middle parameter given the parameters and known data.
NULL data set, I will assume you want all values filled in, for most models with the expected value.NaNs, I will take those as the points to be predicted given the provided data.If the model has no predict method, the default is to use the apop_ml_impute function to do the work. That function does a maximum-likelihood search for the best parameters.
NULL data set, I will return that, with the NaNs filled in. If NULL input, I will allocate an apop_data set and fill it with the expected values.There may be a second page (i.e., a apop_data set attached to the ->more pointer of the main) listing confidence and standard error information. See your specific model documentation for details.
| void apop_prep | ( | apop_data * | d, |
| apop_model * | m | ||
| ) |
Allocate and initialize the parameters, info, and other requisite parts of a apop_model.
Some models have associated prep routines that also attach settings groups to the model, and set up additional special-case functions in vtables.
| int apop_prep_output | ( | char const * | output_name, |
| FILE ** | output_pipe, | ||
| char * | output_type, | ||
| char * | output_append | ||
| ) |
If you're reading this, it is probably because you were referred by another function that uses this internally. You should never call this function directly, but do read this documentation.
There are four settings that affect how output happens, which can be set when you call the function that sent you to this documentation, e.g:
| output_name | The name of the output file, if any. For a database, the table to write. |
| output_pipe | If you have already opened a file and have a FILE* on hand, use this instead of giving the file name. |
| output_type | 'p' = pipe, 'f'= file, 'd' = database |
| output_append | 'a' = append (default), 'w' = write over. |
At the end, output_name, output_pipe, and output_type are all set. Notably, the local output_pipe will have the correct location for the calling function to fprintf to.
| apop_data * apop_rake | ( | char const * | margin_table, |
| char *const * | var_list, | ||
| int | var_ct, | ||
| char *const * | contrasts, | ||
| int | contrast_ct, | ||
| char const * | structural_zeros, | ||
| int | max_iterations, | ||
| double | tolerance, | ||
| char const * | count_col, | ||
| char const * | init_table, | ||
| char const * | init_count_col, | ||
| double | nudge | ||
| ) |
Fit a log-linear model via iterative proportional fitting, aka raking.
Raking has many uses. The Modeling with Data blog presents a series of discussions of uses of raking, including some worked examples.
Or see Wikipedia for an overview of Log linear models, aka Poisson regressions. One approach toward log-linear modeling is a regression form; let there be four categories, A, B, C, and D, from which we can produce a model positing, for example, that cell count is a function of a form like  . In this case, we would assign a separate coefficient to every possible value of A, every possible value of (B, C), and every value of (C, D). Raking is the technique that searches for that large set of parameters.
. In this case, we would assign a separate coefficient to every possible value of A, every possible value of (B, C), and every value of (C, D). Raking is the technique that searches for that large set of parameters.
The combinations of categories that are considered to be relevant are called contrasts, after ANOVA terminology of the 1940s.
The other constraint on the search are structural zeros, which are values that you know can never be non-zero, due to field-specific facts about the variables. For example, U.S. Social Security payments are available only to those age 65 or older, so "age <65 and gets_soc_security=1" is a structural zero.
Because there is one parameter for every combination, there may be millions of parameters to estimate, so the search to find the most likely value requires some attention to technique. For over half a century, the consensus method for searching has been raking, which iteratively draws each category closer to the mean in a somewhat simple manner (this was first developed circa 1940 and had to be feasible by hand), but which is guaranteed to eventually arrive at the maximum likelihood estimate for all cells.
Another complication is that the table is invariably sparse. One can easily construct tables with millions of cells, but the corresponding data set may have only a few thousand observations.
This function uses the database to resolve the sparseness problem. It constructs a query requesting all combinations of categories the could possibly be non-zero after raking, given all of the above constraints. Then, raking is done using only that subset. This means that the work is done on a number of cells proportional to the number of data points, not to the full cross of all categories. Set apop_opts.verbose to 2 or greater to show the query on stderr.
.init_table, then an all-ones default table will be used.| margin_table | The name of the table in the database to use for calculating the margins. The table should have one observation per row. (No default) |
| var_list | The full list of variables to search. A list of strings, e.g., (char *[]){"var1", "var2", ..., "var15"} |
| var_ct | The count of the full list of variables to search. |
| contrasts | The contrasts describing your model. Like the var_list input, a list of strings like (char *[]){"var1", "var7", "var13"} contrast is a pipe-delimited list of variable names. (No default) |
| contrast_ct | The number of contrasts in the list of contrasts. (No default) |
| structural_zeros | a SQL clause indicating combinations that can never take a nonzero value. This will go into a where clause, so anything you could put there is OK, e.g. "age <65 and gets_soc_security=1 or age <15 and married=1". Your margin data is not checked for structural zeros. (default: no structural zeros) |
| max_iterations | Number of rounds of raking at which the algorithm halts. (default: 1000) |
| tolerance | I calculate the change for each cell from round to round; if the largest cell change is smaller than this, I stop. (default: 1e-5) |
| count_col | This column gives the count of how many observations are represented by each row. If NULL, ech row represents one person. (default: NULL) |
| init_table | The default is to initially set all table elements to one and then rake from there. This is effectively the `fully synthetic' approach, which uses only the information in the margins and derives the data set closest to the all-ones data set that is consistent with the margins. Care is taken to maintan sparsity in this case. If you specify an init_table, then I will get the initial cell counts from it. (default: the fully-synthetic approach, using a starting point of an all-ones grid.) |
| init_count_col | The column in init_table with the cell counts. |
| nudge | There is a common hack of adding a small value to every zero entry, because a zero entry will always scale to zero, while a small value could eventually scale to anything. Recall that this function works on sparse sets, so I first filter out those cells that could possibly have a nonzero value given the observations, then I add nudge to any zero cells within that subset. |
weights vector gives the most likely value for each cell.| out->error='i' | Input was somehow wrong. |
| out->error='c' | Raking did not converge, reached max. iteration count. |
apop_opts.verbose=3 to see the intermediate tables at the end of each round of raking. omp_get_thread_num. | int apop_regex | ( | const char * | string, |
| const char * | regex, | ||
| apop_data ** | substrings, | ||
| const char | use_case | ||
| ) |
Extract subsets from a string via regular expressions.
This function takes a regular expression and repeatedly applies it to an input string. It returns the count of matches, and optionally returns the matches themselves organized into the text grid of an apop_data set.
For example, "p.val" will match "P value", "p.value", "p values" (and even "tempeval", so be careful).
If you give a non-NULL address in which to place a table of paren-delimited substrings, I'll return them as a row in the text element of the returned apop_data set. I'll return all the matches, filling the first row with substrings from the first application of your regex, then filling the next row with another set of matches (if any), and so on to the end of the string. Useful when parsing a list of items, for example.
| string | The string to search (no default) |
| regex | The regular expression (no default) |
| substrings | Parens in the regex indicate that I should return matching substrings. Give me the address of an apop_data* set, and I will allocate and fill the text portion with matches. Default= NULL, meaning do not return substrings (even if parens exist in the regex). If no match, return an empty apop_data set, so output->textsize[0]==0. |
| use_case | Should I be case sensitive, 'y' or 'n'? (default = 'n', which is not the POSIX default.) |
substrings may be allocated and filled if needed.apop_opts.stop_on_warning='n' returns -1 on error (e.g., regex NULL or didn't compile). strings==NULL, I return 0—no match—and if substrings is provided, set it to NULL.&subs, not plain subs.([A-Za-z])([0-9]), the column zero of outdata will hold letters, and column one will hold numbers. Use apop_data_transpose to reverse this so that the letters are in outdata->text[0] and numbers in outdata->text[1]. | gsl_rng * apop_rng_alloc | ( | int | seed | ) |
Initialize a gsl_rng.
Uses the Tausworth routine.
| seed | The seed. No need to get funny with it: 0, 1, and 2 will produce wholly different streams. |
| double apop_rng_GHgB3 | ( | gsl_rng * | r, |
| double * | a | ||
| ) |
RNG from a Generalized Hypergeometric type B3.
Devroye uses this as the base for many of his distribution-generators, including the Waring.
| void apop_score | ( | apop_data * | d, |
| gsl_vector * | out, | ||
| apop_model * | m | ||
| ) |
Find the vector of first derivatives (aka the gradient) of the log likelihood of a data/parametrized model pair.
On input, the model m must already be sufficiently prepped that the log likelihood can be evaluated; see p, log_likelihood for details.
On output, the gsl_vector input to the function will be filled with the gradients (or NaNs on errors). If the model parameters have a more complex shape than a simple vector, then the vector will be in apop_data_pack order; use apop_data_unpack to reformat to the preferred shape.
| d | The apop_data set at which the score is being evaluated. |
| out | The score to be returned. I expect you to have allocated this already. |
| m | The parametrized model, which must have either a log_likelihood or a p method. |
| apop_data * apop_t_test | ( | gsl_vector * | a, |
| gsl_vector * | b | ||
| ) |
Answers the question: with what confidence can I say that the means of these two columns of data are different?
If apop_opts.verbose is >=1, then display some information to stdout, like the mean/var/count for both vectors and the t statistic.
| a | one column of data |
| b | another column of data |
mean left - right: the difference in means; if positive, first vector has larger mean, and one-tailed test is testing 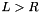 , else reverse if negative.
, else reverse if negative.t statistic: used for the testdf: degrees of freedomp value, 1 tail: the p-value for a one-tailed test that one vector mean is greater than the other.confidence, 1 tail: 1- p value.p value, 2 tail: the p-value for the two-tailed test that left mean = right mean.confidence, 2 tail: 1-p valueExample usage:
| int apop_table_exists | ( | char const * | name, |
| char | remove | ||
| ) |
Check for the existence of a table, and maybe delete it.
Recreating a table which already exists can cause errors, so it is good practice to check for existence first. Also, this is the stylish way to delete a table, since just calling "drop table" will give you an error if the table doesn't exist.
| name | the table name (no default) |
| remove | 'd' ==>delete table so it can be recreated in main. 'n' ==>no action. Return result so program can continue. (default) |
| double apop_test | ( | double | statistic, |
| char * | distribution, | ||
| double | p1, | ||
| double | p2, | ||
| char | tail | ||
| ) |
This is a convenience function to do the lookup of a given statistic along a given distribution. You give me a statistic, its (hypothesized) distribution, and whether to use the upper tail, lower tail, or both. I will return the odds of a Type I error given the model—in statistician jargon, the 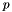 -value. [Type I error: odds of rejecting the null hypothesis when it is true.]
-value. [Type I error: odds of rejecting the null hypothesis when it is true.]
For example,
will return the density of the standard Normal distribution that is more than 1.3 from zero. If this function returns a small value, we can be confident that the statistic is significant. Or,
will give the appropriate odds for an upper-tailed test using the 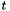 -distribution with 10 degrees of freedom (e.g., a
-distribution with 10 degrees of freedom (e.g., a  -test of the null hypothesis that the statistic is less than or equal to zero).
-test of the null hypothesis that the statistic is less than or equal to zero).
Several more distributions are supported; see below.
| statistic | The scalar value to be tested. |
| distribution | The name of the distribution; see below. |
| p1 | The first parameter for the distribution; see below. |
| p2 | The second parameter for the distribution; see below. |
| tail | 'u' = upper tail; 'l' = lower tail; anything else = two-tailed. (default = two-tailed) |
 -value).
-value).Here are the distributions you can use and their parameters.
"normal" or "gaussian"
 , p2=
, p2= 
"lognormal"
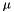 , p2=
, p2= 
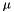 and
and  refer to the Normal one would get after exponentiation
refer to the Normal one would get after exponentiation "uniform"
"t"
"chi squared", "chi", "chisq":
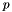 -value for typical cases)
-value for typical cases)"f"
Run a Chi-squared test on an ANOVA table, i.e., an NxN table with the null hypothesis that all cells are equally likely.
| d | The input data, which is a crosstab of various elements. They don't have to sum to one. |
"chi squared statistic", "df", and "p value". Retrieve via, e.g., apop_data_get(out, .rowname="p value"). Run the Fisher exact test on an input contingency table.
| out->error=='p' | Processing error in the test. |
For example:
| apop_data * apop_test_kolmogorov | ( | apop_model * | m1, |
| apop_model * | m2 | ||
| ) |
Run the Kolmogorov-Smirnov test to determine whether two distributions are identical.
| m1 | A sorted PMF model. I.e., a model estimated via something like apop_model *m1 = apop_estimate(apop_data_sort(input_data), apop_pmf); |
| m2 | Another apop_model. If it is a PMF, then I will use a two-sample test, which is different from the one-sample test used if this is not a PMF. |
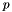 -value from the Kolmogorov-Smirnov test that the two distributions are equal.
-value from the Kolmogorov-Smirnov test that the two distributions are equal.| out->error='m' | Model error: m1 is not an apop_pmf. I verify this by checking whether m1->cdf == apop_pmf->cdf. |
m1 and m2 are identical. A future version of Apophenia may implement a mechanism to allow this function to test for sorted data, but it currently can't.Here is an example, which tests whether a set of draws from a Normal(0, 1) matches a sequence of Normal distributions with increasing mean.
This allocates or resizes the text element of an apop_data set.
If the text element already exists, then this is effectively a realloc function, reshaping to the size you specify.
| in | An apop_data set. It's OK to send in NULL, in which case an apop_data set with NULL matrix and vector elements is returned. |
| row | the number of rows of text. |
| col | the number of columns of text. |
NULL, then this is a repeat of the input pointer. | out->error=='a' | Allocation error. |
| void apop_text_free | ( | char *** | freeme, |
| int | rows, | ||
| int | cols | ||
| ) |
Free a matrix of chars* (i.e., a char***). This is what apop_data_free uses internally to deallocate the text element of an apop_data set. You may never need to use it directly.
Sample usage:
| char * apop_text_paste | ( | apop_data const * | strings, |
| char * | between, | ||
| char * | before, | ||
| char * | after, | ||
| char * | between_cols, | ||
| apop_fn_riip | prune, | ||
| void * | prune_parameter | ||
| ) |
Join together the text grid of an apop_data set into a single string.
For example, say that we have a data set with some text: row 0 has "a0", "b0", "c0"; row 2 has "a1", "b1", "c1"; and so on. We would like to produce
This could be sent to an SQL engine to copy the data to a database (but this is just an example for demonstration—use apop_data_print to write to a database table).
To construct this single string from the text grid, we would need to add:
Insert into tab values ('. ', ' '); \ninsert into tab values(' ');'Thus, do the conversion via:
| strings | An apop_data set with a grid of text to be combined into a single string |
| between | The text to put in between the rows of the table, such as ", ". (Default is a single space: " ") |
| before | The text to put at the head of the string. For the query example, this would be .before="select ". (Default: NULL) |
| after | The text to put at the tail of the string. For the query example, .after=" from data_table". (Default: NULL) |
| between_cols | The text to insert between columns of text. See below for an example (Default is set to equal .between) |
| prune | If you don't want to use the entire text set, you can provide a function to indicate which elements should be pruned out. Some examples: |
| prune_parameter | A void pointer to pass to your prune function. |
strings table joined as per your specification. Allocated by the function, to be freed by you if desired.NULL or has no text, the output string will have only the .before and .after parts with nothing in between. apop_opts.verbose >=3, then print the pasted text to stderr. Apop_r and Apop_rs to get a view of only one or a few rows in conjunction with this function.This sample snippet generates the SQL for a query using a list of column names (where the query begins with select , ends with from datatab, and has commas in between each element), re-processes the same list to produce the head of an HTML table, then produces the body of the table with the query result.
| int apop_text_set | ( | apop_data * | in, |
| const size_t | row, | ||
| const size_t | col, | ||
| const char * | fmt, | ||
| ... | |||
| ) |
Add a string to the text element of an apop_data set. If you send me a NULL string, I will write the value of apop_opts.nan_string in the given slot. If there is already something in that slot, that string is freed, preventing memory leaks.
| in | The apop_data set, that already has an allocated text element. |
| row | The row |
| col | The column |
| fmt | The text to write. |
| ... | You can use a printf-style fmt and follow it with the usual variables to fill in. |
asprintf), not a pointer to the input(s). NULL, write apop_opts.nan_string at that point. You may prefer to use "" to express a blank. | apop_data * apop_text_to_data | ( | char const * | text_file, |
| int | has_row_names, | ||
| int | has_col_names, | ||
| int const * | field_ends, | ||
| char const * | delimiters | ||
| ) |
Read a delimited or fixed-wisdth text file into the matrix element of an apop_data set.
See Input text file formatting.
See also apop_text_to_db, which handles text data, and may othewise be a perferable approach to data management.
| text_file | = "-" The name of the text file to be read in. If "-" (the default), use stdin. |
| has_row_names | Does the lines of data have row names? 'y' =yes; 'n' =no (default: 'n') |
| has_col_names | Is the top line a list of column names? See Input text file formatting for notes on dimension (default: 'y') |
| field_ends | If fields have a fixed size, give the end of each field, e.g. .field_ends=(int[]){3, 8 11}. (default: NULL, indicating not fixed width) |
| delimiters | A string listing the characters that delimit fields. (default: "|,\t") |
| out->error=='a' | allocation error |
| out->error=='t' | text-reading error |
example: See apop_ols.
| int apop_text_to_db | ( | char const * | text_file, |
| char * | tabname, | ||
| int | has_row_names, | ||
| int | has_col_names, | ||
| char ** | field_names, | ||
| int const * | field_ends, | ||
| apop_data * | field_params, | ||
| char * | table_params, | ||
| char const * | delimiters, | ||
| char | if_table_exists | ||
| ) |
Read a delimited or fixed-width text file into a database table. See Input text file formatting.
For purely numeric data, you may be able to bypass the database by using apop_text_to_data.
See the apop_ols page for an example that uses this function to read in sample data (also listed on that page).
Apophenia ships with an apop_text_to_db command-line utility, which is a wrapper for this function.
Especially if you are using a pre-2007 version of SQLite, there may be a speedup to putting this function in a begin/commit wrapper:
| text_file | The name of the text file to be read in. If "-", then read from STDIN. (default: "-") |
| tabname | The name to give the table in the database (default: text_file after the last slash and up to the next dot. E.g., text_file=="../data/pant_lengths.csv" gives tabname=="pant_lengths") |
| has_row_names | Does the lines of data have row names? (default: 0) |
| has_col_names | Is the top line a list of column names? (default: 1) |
| field_names | The list of field names, which will be the columns for the table. If has_col_names==1, read the names from the file (and just set this to NULL). If has_col_names == 1 && field_names !=NULL, I'll use the field names. (default: NULL) |
| field_ends | If fields have a fixed size, give the end of each field, e.g. .field_ends=(int[]){3, 8 11}. (default: NULL, indicating not fixed width) |
| field_params | There is an implicit create table in setting up the database. If you want to add a type, constraint, or key, put that here. The relevant part of the input apop_data set is the text grid, which should be  . The first item in each row ( . The first item in each row (your_params->text[n][0], for each 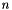 ) is a regular expression to match against the variable names; the second item ( ) is a regular expression to match against the variable names; the second item (your_params->text[n][1]) is the type, constraint, and/or key (i.e., what comes after the name in the create query). Not all variables need be mentioned; the default type if nothing matches is numeric. I go in order until I find a regex that matches the given field, so if you don't like the default, then set the last row to have name .*, which is a regex guaranteed to match anything that wasn't matched by an earlier row, and then set the associated type to your preferred default. See apop_regex on details of matching. (default: NULL) |
| table_params | There is an implicit create table in setting up the database. If you want to add a table constraint or key, such as not null primary key (age, sex), put that here. |
| delimiters | A string listing the characters that delimit fields. default = "|,\t" |
| if_table_exists | What should I do if the table exists?'n' Do nothing; exit this function. (default)'d' Retain the table but delete all data; refill with the new data (i.e., call "delete * from your_table").'o' Overwrite the table from scratch; deleting the previous table entirely.'a' Append new data to the existing table. |
Give me a column of text, and I'll give you a sorted list of the unique elements. This is basically running select distinct * from datacolumn, but without the aid of the database.
| d | An apop_data set with a text component |
| col | The text column you want me to use. |
| apop_model * apop_update | ( | apop_data * | data, |
| apop_model * | prior, | ||
| apop_model * | likelihood, | ||
| gsl_rng * | rng | ||
| ) |
Take in a prior and likelihood distribution, and output a posterior distribution.
p or log_likelihood element, then use apop_model_metropolis to generate the posterior. If you expect MCMC to run, you may add an apop_mcmc_settings group to your prior to control the details of the search. See also the apop_model_metropolis documentation.p or log_likelihood but does have a draw element, then make draws from the prior and weight them by the p given by the likelihood distribution. This is not a rejection sampling method, so the burnin is ignored.| data | The input data, that will be used by the likelihood function (default = NULL.) |
| prior | The prior apop_model. If the system needs to estimate the posterior via MCMC, this needs to have a log_likelihood or p method. (No default, must not be NULL.) |
| likelihood | The likelihood apop_model. If the system needs to estimate the posterior via MCMC, this needs to have a log_likelihood or p method (ll preferred). (No default, must not be NULL.) |
| rng | A gsl_rng, already initialized (e.g., via apop_rng_alloc). (default: an RNG from apop_rng_get_thread) |
| Prior | Likelihood | Notes |
| Beta | Binomial | |
| Beta | Bernoulli | |
| Exponential | Gamma | Gamma likelihood represents the distribution of  , not plain , not plain  |
| Normal | Normal | Assumes prior with fixed  ; updates distribution for ; updates distribution for 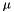 |
| Gamma | Poisson | Uses sum and size of the data |
Here is a test function that compares the output via conjugate table and via Metropolis-Hastings sampling:
| void apop_vector_apply | ( | gsl_vector * | v, |
| void(*)(double *) | fn | ||
| ) |
Apply a function to every row of a matrix. The function that you input takes in a double* and may modify the input value in place. This function will send a pointer to each element of your vector to your function.
| v | The input vector |
| fn | A function of the form void fn(double in) |
NULL, this is a no-op. | int apop_vector_bounded | ( | const gsl_vector * | in, |
| long double | max | ||
| ) |
Test that all elements of a vector are within bounds, so you can preempt a procedure that is about to break on infinite or too-large values.
| in | A gsl_vector |
| max | An upper and lower bound to the elements of the vector. (default: INFINITY) |
 ;
;NULL vector has no unbounded elements, so NULL input returns 1. You get a warning if apop_opts.verbosity >=2. | gsl_vector * apop_vector_copy | ( | const gsl_vector * | in | ) |
Copy one gsl_vector to another. That is, all data is duplicated. Unlike gsl_vector_memcpy, this function allocates and returns the destination, so you can use it like this:
| in | The input vector |
gsl_vector_alloc fails, returns NULL and print a warning. | double apop_vector_correlation | ( | const gsl_vector * | ina, |
| const gsl_vector * | inb, | ||
| const gsl_vector * | weights | ||
| ) |
Returns the correlation coefficient of two vectors: 
An example
| ina,inb | Two vectors of equal length (no default, must not be NULL) |
| weights | Replicate weights for the observations. (default: equal weights for all observations) |
| double apop_vector_cov | ( | const gsl_vector * | v1, |
| const gsl_vector * | v2, | ||
| const gsl_vector * | weights | ||
| ) |
Find the sample covariance of a pair of vectors, with an optional weighting. This only makes sense if the weightings are identical, so the function takes only one weighting vector for both.
| v1,v2 | The data vectors (no default; must not be NULL) |
| weights | The weight vector. (default equal weights for all elements) |
| double apop_vector_distance | ( | const gsl_vector * | ina, |
| const gsl_vector * | inb, | ||
| const char | metric, | ||
| const double | norm | ||
| ) |
Returns the distance between two vectors, where distance is defined based on the third (optional) parameter:
 where
where 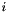 iterates over dimensions.
iterates over dimensions.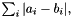 where
where 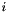 iterates over dimensions.
iterates over dimensions.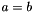 , return zero, else return one.
, return zero, else return one.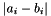 is largest, return the distance along that one dimension.
is largest, return the distance along that one dimension.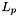 norm,
norm,  . The value of
. The value of 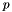 is set by the fourth (optional) argument.
is set by the fourth (optional) argument.| ina | First vector (No default, must not be NULL) |
| inb | Second vector (Default = zero) |
| metric | The type of metric, as above. |
| norm | If you are using an 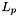 norm, this is norm, this is 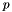 . Must be strictly greater than zero. (default = 2) . Must be strictly greater than zero. (default = 2) |
v, its longest element, and its sum. | long double apop_vector_entropy | ( | gsl_vector * | in | ) |
Given a vector representing a probability distribution of observations, calculate the entropy,  .
.
 ; see the example.
; see the example.NULL or a total weight of zero) is zero. Print a warning when given NULL input and apop_opts.verbose >=1.NaN; print a warning when apop_opts.verbose >= 0.Sample code:
| void apop_vector_exp | ( | gsl_vector * | v | ) |
Replace every vector element 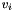 with exp
with exp  .
.
NULL, do nothing. | double apop_vector_kurtosis | ( | const gsl_vector * | in | ) |
Returns the sample fourth central moment of the data in the given vector. Corrections are made to produce an unbiased result as per Appendix M (PDF) of Modeling with data.
 is
is  , not three, one, or zero.
, not three, one, or zero. | double apop_vector_kurtosis_pop | ( | gsl_vector const * | v, |
| gsl_vector const * | weights | ||
| ) |
Returns the population fourth central moment [  ] of the data in the given vector, with an optional weighting.
] of the data in the given vector, with an optional weighting.
| v | The data vector |
| weights | The weight vector. If NULL, assume equal weights. |
| void apop_vector_log | ( | gsl_vector * | v | ) |
Replace every vector element  with ln
with ln  .
.
NULL, do nothing. | void apop_vector_log10 | ( | gsl_vector * | v | ) |
Replace every vector element  with log
with log 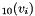 .
.
NULL, do nothing. | gsl_vector * apop_vector_map | ( | const gsl_vector * | v, |
| double(*)(double) | fn | ||
| ) |
Map a function onto every element of a vector. Thus function will send each element to the function you provide, and will output a gsl_vector holding your function's output for each row.
| v | The input vector |
| fn | A function of the form double fn(double in) |
gsl_vector (allocated by this function) with the corresponding value for each row.NULL vector, I return NULL. | double apop_vector_map_sum | ( | const gsl_vector * | in, |
| double(*)(double) | fn | ||
| ) |
Returns the sum of the output of apop_vector_map. For example, apop_vector_map_sum(v, isnan) returns the count of elements of v that are NaN.
NULL vector, I return the sum of zero items: zero. | double apop_vector_mean | ( | gsl_vector const * | v, |
| gsl_vector const * | weights | ||
| ) |
Find the mean, weighted or unweighted.
| v | The data vector |
| weights | The weight vector. Default: assume equal weights. |
| int gsl_vector * apop_vector_moving_average | ( | gsl_vector * | v, |
| size_t | bandwidth | ||
| ) |
Return a new vector that is the moving average of the input vector.
| v | The input vector, unsmoothed |
| bandwidth | An integer  giving the number of elements to be averaged to produce one number. giving the number of elements to be averaged to produce one number. |
v->size - (bandwidth/2)*2. | void apop_vector_normalize | ( | gsl_vector * | in, |
| gsl_vector ** | out, | ||
| const char | normalization_type | ||
| ) |
This function will normalize a vector, either such that it has mean zero and variance one, or ranges between zero and one, or sums to one.
| in | A gsl_vector with the un-normalized data. NULL input gives NULL output. (No default) |
| out | If normalizing in place, NULL. If not, the address of a gsl_vector*. Do not allocate. (default = NULL.) |
| normalization_type | 'p': normalized vector will sum to one. E.g., start with a set of observations in bins, end with the percentage of observations in each bin. (the default)'r': normalized vector will range between zero and one. Replace each X with (X-min) / (max - min).'s': normalized vector will have mean zero and (sample) variance one. Replace each X with 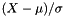 , where , where  is the sample standard deviation. is the sample standard deviation.'m': normalize to mean zero: Replace each X with  |
Example
| double * apop_vector_percentiles | ( | gsl_vector * | data, |
| char | rounding | ||
| ) |
Returns an array of size 101, where returned_vector[95] gives the value of the 95th percentile, for example. Returned_vector[100] is always the maximum value, and returned_vector[0] is always the min (regardless of rounding rule).
| data | A gsl_vector with the data. (No default, must not be NULL.) |
| rounding | Either be 'u', 'd', or 'a'. Unless your data is exactly a multiple of 101, some percentiles will be ambiguous. If 'u', then round up (use the next highest value); if 'd', round down to the next lowest value; if 'a', take the mean of the two nearest points. (Default = 'd'.) |
'u' or 'a', then you can say "5% or more of
the sample is below returned_vector[5]"; if 'd' or 'a', then you can say "5%
or more of the sample is above returned_vector[5]". free() the array returned by this function. | void apop_vector_print | ( | gsl_vector * | data, |
| Output_declares | |||
| ) |
Print a vector to the screen, a file, a pipe, or the database.
apop_opts.output_type="\n" would print the vector vertically. | gsl_vector * apop_vector_realloc | ( | gsl_vector * | v, |
| size_t | newheight | ||
| ) |
This function will resize a gsl_vector to a new length.
Data in the vector will be retained. If the new height is smaller than the old, then data at the end of the vector will be cropped away (in a non–memory-leaking manner). If the new height is larger than the old, then new cells will be filled with garbage; it is your responsibility to zero out or otherwise fill them before use.
reallocs can take a noticeable amount of time. You are thus encouraged to make an effort to determine the size of your data and do one allocation, rather than writing for loops that resize a vector at every increment. gsl_vector is a versatile struct that can represent subvectors, matrix columns and other cuts from parent data. Resizing a portion of a parent matrix makes no sense, so return NULL and print an error if asked to resize a view.| v | The already-allocated vector to resize. If you give me NULL, this is equivalent to gsl_vector_alloc |
| newheight | The height you'd like the vector to be. |
| double apop_vector_skew | ( | const gsl_vector * | in | ) |
Returns an unbiased estimate of the sample skew of the data in the given vector.
| double apop_vector_skew_pop | ( | gsl_vector const * | v, |
| gsl_vector const * | weights | ||
| ) |
Returns the population skew 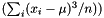 of the data in the given vector. Observations may be weighted.
of the data in the given vector. Observations may be weighted.
| v | The data vector |
| weights | The weight vector. Default: equal weights for all observations. |
 ; that's not done here, so you'll have to do so separately if need be.
; that's not done here, so you'll have to do so separately if need be.w->size as the number of elements, and returns the usual sum over  . If weights > 1, then the system uses the total weights as
. If weights > 1, then the system uses the total weights as 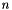 . Thus, you can use the weights as standard weightings or to represent elements that appear repeatedly.
. Thus, you can use the weights as standard weightings or to represent elements that appear repeatedly. | gsl_vector * apop_vector_stack | ( | gsl_vector * | v1, |
| gsl_vector const * | v2, | ||
| char | inplace | ||
| ) |
Put the first vector on top of the second vector.
| v1 | the upper vector (default=NULL, in which case this copies v2) |
| v2 | the second vector (default=NULL, in which case nothing is added) |
| inplace | If 'y', use apop_vector_realloc to modify v1 in place; see the caveats on that function. Otherwise, allocate a new vector, leaving v1 undisturbed. (default='n') |
v1.| long double apop_vector_sum | ( | const gsl_vector * | in | ) |
Returns the sum of the data in the given vector.
| gsl_matrix * apop_vector_to_matrix | ( | const gsl_vector * | in, |
| char | row_col | ||
| ) |
This function copies the data in a vector to a new one-column (or one-row) matrix and returns the newly-allocated and filled matrix.
For the reverse, try apop_data_pack.
| in | a gsl_vector (No default. If NULL, I return NULL, with a warning if apop_opts.verbose >=1 ) |
| row_col | If 'r', then this will be a row (1 x N) instead of the default, a column (N x 1). (default: 'c') |
gsl_matrix with one column (or row).NULL vector, you get a NULL pointer in return. I warn you of this if apop_opts.verbosity >=2 . gsl_matrix_alloc fails you get a NULL pointer in return. | gsl_vector * apop_vector_unique_elements | ( | const gsl_vector * | v | ) |
Give me a vector of numbers, and I'll give you a sorted list of the unique elements. This is basically running select distinct datacol from data order by datacol, but without the aid of the database.
| v | a vector of items |
| double apop_vector_var | ( | gsl_vector const * | v, |
| gsl_vector const * | weights | ||
| ) |
Find the sample variance of a vector, weighted or unweighted.
| v | The data vector |
| weights | The weight vector. If NULL (the default), assume equal weights. |
 instead of
instead of 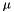 .
. w->size as the number of elements, and returns the usual sum over  . If weights > 1, then the system uses the total weights as
. If weights > 1, then the system uses the total weights as 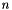 . Thus, you can use the weights as standard weightings or to represent elements that appear repeatedly.
. Thus, you can use the weights as standard weightings or to represent elements that appear repeatedly. | double apop_vector_var_m | ( | const gsl_vector * | in, |
| const double | mean | ||
| ) |
Returns the variance of the data in the given vector, given that you've already calculated the mean.
| in | the vector in question |
| mean | the mean, which you've already calculated using apop_vector_mean. |
| apop_opts_type apop_opts |
Here are where the options are initially set. See the apop_opts_type documentation for details.
| apop_opts_type apop_opts |
Here are where the options are initially set. See the apop_opts_type documentation for details.
| apop_opts_type apop_opts |
Here are where the options are initially set. See the apop_opts_type documentation for details.
| apop_opts_type apop_opts |
Here are where the options are initially set. See the apop_opts_type documentation for details.
| apop_opts_type apop_opts |
Here are where the options are initially set. See the apop_opts_type documentation for details.