The apop_model is intended to provide a consistent expression of any model that (implicitly or explicitly) expresses a likelihood of data given parameters, including traditional linear models, textbook distributions, Bayesian hierarchies, microsimulations, and any combination of the above. The unifying feature is that all of the models act over some data space and some parameter space (in some cases one or both is the empty set), and can assign a likelihood for a fixed pair of parameters and data given the model. This is a very broad requirement, often used in the statistical literature. For discussion of the theoretical structures, see A Useful Algebraic System of Statistical Models (PDF).
This page is about writing new models from scratch, beginning with basic models and on up to models with arbitrary internal settings, specific methods of Bayesian updating using your model as a prior or likelihood, and so on. I assume you have already read Models on using models and have tried a few things with the canned models that come with Apophenia, so you already know how a user handles basic estimation, adding a settings group, and so on.
This page includes:
Users are encouraged to always use models via the helper functions, like apop_estimate or apop_cdf. The helper functions do some boilerplate error checking, and call defaults as needed. For example, if your model has a log_likelihood method but no p method, then apop_p will use exp(log_likelihood). If you don't give an estimate method, then apop_estimate will call apop_maximum_likelihood.
So the game in writing a new model is to write just enough internal methods to give the helper functions what they need. In the not-uncommon best case, all you need to do is write a log likelihood function or an RNG.
Here is how one would set up a model that could be estimated using maximum likelihood:
data is the input data, and m is the parametrized model (i.e. your model with a parameters element already filled in by the caller). This function will return the value of the log likelihood function at the given parameters..name, a human-language name for your model. vsize, msize1, and msize2 elements specify the shape of the parameter set. For example, if there are three numbers in the vector, then set .vsize=3 and omit the matrix sizes. The default model prep routine will call new_est->parameters = apop_data_alloc(vsize, msize1, msize2). dsize element is the size of one random draw from your model. -1 for vsize, msize(1|2), or dsize. If the allocation is exceptional in a different way, then you will need to allocate parameters by writing a custom prep method for the model. .constraint element for those too. See Setting Constraints for more.You already have more than enough that something like this will work (the dsize is used for random draws):
Once that baseline works, you can fill in other elements of the apop_model as needed.
For example, if you are using a maximum likelihood method to estimate parameters, you can get much faster estimates and better covariance estimates by specifying the dlog likelihood function (aka the score):
The score is not part of the model object, but is registered (see below) using
Many procedures in Apophenia use OpenMP to thread operations, so assume your functions are running in a threaded environment. If a method can not be threaded, wrap it in an OpenMP critical region. E.g.,
Your model may need additional settings or auxiliary information to function, which would require associating a model-specific struct with the model. A method associated with a model that uses such a struct usually begins with calls like
These model-specific structs are handled as expected by apop_model_copy and apop_model_free, and many functions that modify or transform an apop_model try to handle settings groups as expected. This section describes how to build a settings group so all these automatic steps happen as expected, and your methods can reliably retrieve settings as needed.
But before getting into the detail of how to make model-specific groups of settings work, note that there's a lightweight method of storing sundry settings, so in many cases you can bypass all of the following. The apop_model structure has a void pointer named more which you can use to point to a model-specific struct. If more_size is larger than zero (i.e. you set it to your_model.more_size=sizeof(your_struct)), then it will be copied via memcpy by apop_model_copy, and freed by apop_model_free. Apophenia's routines will never impinge on this item, so do what you wish with it.
The remainder of this subsection describes the information you'll have to provide to make use of the conveniences described to this point: initialization of defaults, smarter copying and freeing, and adding to an arbitrarily long list of settings groups attached to a model. You will need four items: a typedef for the structure itself, plus init, copy, and free functions. This is the sort of boilerplate that will be familiar to users of object-oriented languages in the style of C++ or Java, but it's really a list of arbitrarily-typed elements, which makes this feel more like LISP. [And being a reimplementation of an existing feature of LISP, this section will be macro-heavy.]
The settings struct will likely go into a header file, so here is a sample header for a new settings group named ysg_settings, with a dataset, two sizes, and a reference counter. ysg stands for Your Settings Group; replace that substring with your preferred name in every instance to follow.
The first item is a familiar structure definition. The last line is a macro that declares the init, copy, and free functions discussed below. This is everything you would need in a header file, should you need one. These are just declarations; we'll write the actual init/copy/free functions below.
The structure itself gets the full name, ysg_settings. Everything else is a macro keyed on ysg, without the _settings part. Because of these macros, your struct name must end in _settings.
If you have an especially simple structure, then you can generate the three functions with these three macros in your .c file:
These macros generate appropriate functions to do what you'd expect: allocating the main structure, copying one struct to another, freeing the main structure. The spaces after the commas indicate that in these cases no special code gets added to the functions that these macros expand into.
You'll never call the generated functions directly; they are called by Apop_settings_add_group, apop_model_free, and other model or settings-group handling functions.
Now that initializing/copying/freeing of the structure itself is handled, the remainder of this section will be about how to add instructions for the structure internals, like data that is pointed to by the structure elements.
NULL. Otherwise, you will need a new line declaring a default for every element in your structure. There is a macro to help with this too. These macros will define for your use a structure named in, and an output pointer-to-struct named out. Continuing the above example: Apop_settings_add(a_model, ysg, .size1=100) would set up a group with a 100-by-10 data set, and set the reference counter allocated and to one.in for your use. Continuing the example: With those three macros in place and the header as above, Apophenia will treat your settings group like any other, and users can use Apop_settings_add_group to populate it and attach it to any model.
The settings groups are for adding arbitrary model-specific nouns; vtables are for adding arbitrary model-specific verbs.
Many functions (e.g., entropy, the dlog likelihood, Bayesian updating) have special cases for well-known models like the Normal distribution. Any function may maintain a registry of models and associated special-case procedures, aka a vtable.
Lookups happen based on a hash that takes into account the elements of the model that will be used in the calculation. For example, the apop_update_hash takes in two models and calculates the hash based on the address of the prior's draw method and the likelihood's log_likelihood or p method. Thus, a vtable lookup for new models that re-use the same methods (at the same addresses in memory) will still find the same special-case function.
If you need to deregister the function, use the associated deregister function, e.g. apop_update_vtable_drop(apop_beta, apop_binomial). You can guarantee that a method will not be re-added by following up the _drop with, e.g., apop_update_vtable_add(NULL, apop_beta, apop_binomial).
The steps for adding a function to an existing vtable:
_vtable_add function to add the function and associate it with the given model. For example, to add a Beta-binomial routine named betabinom to the registry of Bayesian updating routines, use apop_update_vtable_add(betabinom, apop_beta, apop_binomial). ..._vtable_add in the prep method of the given model, thus ensuring that the auxiliary functions are registered after the first time the model is sent to apop_estimate.The easiest way to set up a new vtable is to copy/paste/modify an existing one. Briefly:
apop.h. draw, log_likelihood, and methods of the model. A model where these elements are identical will still match even if other elements are different.The remainder of this section covers the detailed expectations regarding the elements of the apop_model structure. I begin with the data (non-function) elements, and then cover the method (function) elements. Some of the following will be requirements for all models and some will be advice to authors; I use the accepted definitions of "must", "shall", "may" and related words.
data element is treated as a single observation by many functions. For example, apop_bootstrap_cov depends on each row being an iid observation to function correctly. Calculating the Bayesian Information Criterion (BIC) requires knowing the number of observations in the data, and assumes that row count==observation count. For complex data, the apop_data_pack and apop_data_unpack functions can help with this.->matrix element of the apop_data set sent to model methods. Your likelihood, p, cdf, and estimate routines must accept data as a single row of the matrix of the apop_data set for such functions to work. They may accept other formats. Tip: you can use apop_data_pack and apop_data_unpack to convert a structured set to a single row and back again.ols_shuffle to convert a matrix where the first column is the dependent variable to a data set with dependent variable in the vector and a column of ones in the first matrix column.prep method of the model; see below. Given the model m and its elements m.vsize, m.msize1, m.msize2, functions that need to allocate a parameter set will do so via apop_data_alloc(m.vsize, m.msize1, m.msize2).<info>, is typically a list of scalars. Nothing is guaranteed, but the elements may include: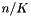 is small (say
is small (say 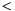 40)'' [Kenneth P. Burnham, David R. Anderson: Model Selection and Multi-Model Inference, p 66, emphasis in original.]
40)'' [Kenneth P. Burnham, David R. Anderson: Model Selection and Multi-Model Inference, p 66, emphasis in original.] For those elements that require a count of input data, the calculations assume each row in the input apop_data set is a single datum.
Get these via, e.g., apop_data_get(your_model->info, .rowname="log likelihood"). When writing for any arbitrary function, be prepared to handle NaN, indicating that the element is not calculated or saved in the info page by the given model.
For OLS-type estimations, each row corresponds to the row in the original data. For filling in of missing data, the elements may appear anywhere, so the row/col indices are essential.
In object-oriented jargon, settings groups are the private elements of the data set, to be pulled out in certain contexts, and ignored in all others. Therefore, there are no rules about internal use. The more element of the apop_model provides a lightweight means of attaching an arbitrary struct to a model. See Writing new settings groups above for details.
more pointer points to a structure or value (let it be ss), then more_size must be set to sizeof(ss).long double your_p_or_ll(apop_data *d, apop_model *params). ->parameters element, possibly a settings group added by the user). NULL and set the model's error element to 'p' if they are missing. NaN on errors. If an error in the input model is found, the function may set the input model's error element to an appropriate char value. log_likelihood and p methods, it must be the case that log(p(d, m)) equals log_likelihood(d, m) for all d and m. This implies that p must return a value 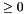 . Note that apop_maximum_likelihood will accept functions where
. Note that apop_maximum_likelihood will accept functions where p returns a negative value, but diagonstics that depend on log likelihood like AIC will return NaN. void your_prep(apop_data *data, apop_model *params). data pointer shall be set to point to the input data. info element shall be allocated and its title set to <Info>. vsize, msize1, or msize2 are -1, then the prep function shall set them to the width of the input data. dsize is -1, then the prep function shall set it to the width of the input data. parameters element is not allocated, the function shall allocate it via apop_data_alloc(vsize, msize1, msize2) (or equivalent). data, and the apop_pmf prep routine calls apop_data_pmf_compress on the input data. void your_estimate(apop_data *data, apop_model *params). It modifies the input model, and returns nothing. Note that this is different from the wrapper function, apop_estimate, which makes a copy of its input model, preps it, and then calls the estimate function with the prepeped copy. parameters hold garbage (as in a malloc without a subsequent assignment to the malloc-ed space). parameters of the input model. For consistency with other models, the estimate should be the maximum likelihood estimate, unless otherwise documented. <Info> page may be filled with statistics, as discussed at infosubsec. For scalars like log likelihood and AIC, use apop_data_add_named_elmt. estimate routine; any changes to the data made by estimate must be documented. error element to a single character. Documentation should include the list of error characters and their meaning.void your_draw(double out, gsl_rng r, apop_model *params) paramters are set, via apop_estimate or apop_model_set_parameters. The author of the draw method should check that parameters are not NULL if needed and fill the output with NaNs if necessary parameters are not set. double of length dsize; user is expected to make sure that there is adequate space. Caller also inputs a gsl_rng, already allocated (probably via apop_rng_alloc, possibly from apop_rng_get_thread). p method. Data shall be reduced to a single vector via apop_data_pack if it is not already a single vector.long double your_cdf(apop_data *d, apop_model *params). parameters are set, via apop_estimate or apop_model_set_parameters. The author of the CDF method should check that parameters are not NULL and return NaN if necessary parameters are not set. matrix of the input apop_data set (as per a draw produced using the draw method). May accept other formats. 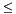 the first row of the input data. The definition of
the first row of the input data. The definition of 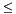 is chosen by the model author.
is chosen by the model author. apop_cdf_settings group may be added to the model to store temp data. See the apop_cdf function for details.long double your_constraint(apop_data *data, apop_model *params). parameters are set, via apop_estimate, apop_model_set_parameters, or the internals of an MLE search. The author of the constraint method should check that parameters are not NULL and return NaN if necessary parameters are not set. parameters in the input model to a constraint-satisfying value, and (2) return the distance between the input parameters and what you've moved the parameters to. The choice of within-bounds parameters and distance function is left to the author of the constraint function.