This is a "gentle introduction" to the Apophenia library. It is intended to give you some initial bearings on the typical workflow and the concepts and tricks that the manual pages assume you are familiar with.
If you want to install Apophenia now so you can try the samples on this page, see the Setting up page.
An outline of this overview:
The opening example
Setting aside the more advanced applications and model-building tasks, let us begin with the workflow of a typical fitting-a-model project using Apophenia's tools:
estimate step runs a few for you), or send the model's output to be input to another model.Here is an example of most of the above steps which you can compile and run, to be discussed in detail below.
The program relies on the U.S. Census's American Community Survey public use microdata for DC 2008, which you can get from the command line via:
or by pointing your browser to that address and saving the file.
The program:
If you saved the code to census.c and don't have a Makefile or other build system, then you can compile it with
or
and then run it with ./census. This compile line will work on any system with all the requisite tools, but for full-time work with this or any other C library, you will probably want to write a Makefile.
The results are unremarkable—age has a positive effect on income, and sex (1=male, 2=female) does has a negative effect—but it does give us some lines of code to dissect.
The first two lines in main() make use of a database. I'll discuss the value of the database step more at the end of this page, but for now, note that there are several functions, apop_query and apop_query_to_data being the ones you will most frequently be using, that will allow you to talk to and pull data from either an SQLite or mySQL/mariaDB database. The database is a natural place to do data processing like renaming variables, selecting subsets, and transforming values.
Designated initializers
Like this line,
many Apophenia functions accept named, optional arguments. To give another example, the apop_data set has the usual row and column numbers, but also row and column names. So you should be able to refer to a cell by any combination of name or number; for the data set you read in above, which has column names, all of the following work:
Default values mean that the apop_data_get, apop_data_set, and apop_data_ptr functions handle matrices, vectors, and scalars sensibly:
These conveniences may be new to users of less user-friendly C libraries, but it it fully conforms to the C standard (ISO/IEC 9899:2011). See the Designated initializers page for details.
A lot of real-world data processing is about quotidian annoyances about text versus numeric data or dealing with missing values, and the apop_data set and its many support functions are intended to make data processing in C easy. Some users of Apophenia use the library only for its apop_data set and associated functions. See Data sets for extensive notes on using the structure.
The structure includes seven parts:
This is not a generic and abstract ideal, but is the sort of mess that real-world data sets look like. For example, here is some data for a weighted OLS regression. It includes an outcome variable in the vector, dependent variables in the matrix and text grid, replicate weights, and column names in bold labeling the variables:
| Rowname | Vector | Matrix | Text | Weights | ||||||||||||||||||||||||||||||||||
|
|
|
|
|
Apophenia's functions generally assume that one row across all of these elements describes a single observation or data point.
See above for some examples of getting and setting individual elements.
Also, apop_data_get, apop_data_set, and apop_data_ptr consider the vector to be the -1st column, so using the data set in the figure, apop_data_get(sample_set, .row=0, .col=-1) == 1.
Reading in data
As per the example above, use apop_text_to_data or apop_text_to_db and then apop_query_to_data.
Subsets
There are many macros to get views of subsets of the data. Each generates a disposable wrapper around the base data: once the variable goes out of scope, the wrapper disappears, but modifications made to the data in the view are modifications to the base data itself.
All of these slicing routines are macros, because they generate several background variables in the current scope (something a function can't do). Traditional custom is to put macro names in all caps, like APOP_DATA_ROWS, which to modern sensibilities looks like yelling. The custom has a logic: there are ways to hang yourself with macros, so it is worth distinguishing them typographically. Apophenia tones it down by capitalizing only the first letter.
Basic manipulations
See Data sets for a list of many other manipulations of data sets, such as apop_data_listwise_delete for quick-and-dirty removal of observations with NaNs, apop_data_split / apop_data_stack, or apop_data_sort to sort all elements by a single column.
Apply and map
If you have an operation of the form for each element of my data set, call this function, then you can use apop_map to do it. You could basically do everything you can do with an apply/map function via a for loop, but the apply/map approach is clearer and more fun. Also, if you set OpenMP's omp_set_num_threads(N) for any N greater than 1 (the default on most systems is the number of CPU cores), then the work of mapping will be split across multiple CPU threads. See Map/apply for a number of examples.
Text
String handling in C usually requires some tedious pointer and memory handling, but the functions to put strings into the text grid in the apop_data structure and get them out again will do the pointer shunting for you. The apop_text_alloc function is really a realloc function: you can use it to resize the text grid as necessary. The apop_text_set function will write a single string to the grid, though you may be using apop_query_to_text or apop_query_to_mixed_data to read in an entire data set at once. Functions that act on entire data sets, like apop_data_rm_rows, handle the text part as well.
The text grid for your_data has your_data->textsize[0] rows and your_data->textsize[1] columns. If you are using only the functions to this point, then empty elements are a blank string (""), not NULL. For reading individual elements, refer to the  th text element via
th text element via your_data->text[i][j].
Errors
Many functions will set the error element of the apop_data structure being operated on if anything goes wrong. You can use this to halt the program or take corrective action:
The whole structure
Here is a diagram of all of Apophenia's structures and how they relate. It is taken from this cheat sheet on general C and SQL use (2 page PDF).

All of the elements of the apop_data structure are laid out at middle-left. You have already met the vector, matrix, weights, and text grid.
The diagram shows the apop_name structure, which has received little mention so far because names basically take care of themselves. A query will bring in column names (and row names if you set apop_opts.db_name_column), or use apop_data_add_names to add names to your data set and apop_name_stack to copy from one data set to another.
The apop_data structure has a more element, for when your data is best expressed in more than one page of data. Use apop_data_add_page, apop_data_rm_page, and apop_data_get_page. Output routines will sometimes append an extra page of auxiliary information to a data set, such as pages named <Covariance> or <Factors>. The angle-brackets indicate a page that describes the data set but is not a part of it (so an MLE search would ignore that page, for example).
Now let us move up the structure diagram to the apop_model structure.
Even restricting ourselves to the most basic operations, there are a lot of things that we want to do with our models: use a data set to estimate the parameters of a model (like the mean and variance of a Normal distribution), or draw random numbers, or show the expected value, or show the expected value of one part of the data given fixed values for the rest of it. The apop_model is intended to encapsulate most of these desires into one object, so that models can easily be swapped around, modified to create new models, compared, and so on.
From the figure above, you can see that the apop_model structure includes a number of informational items, key being the parameters, data, and info elements; a list of settings to be discussed below; and a set of procedures for many operations. Its contents are not (entirely) arbitrary: the overall intent and the theoretical basis for what is and is not included in an apop_model are described in this U.S. Census Bureau research report.
There are helper functions that will allow you to avoid dealing with the model internals. For example, the apop_estimate helper function means you never have to look at the model's estimate method (if it even has one), and you will simply pass the model to a function, as with the above form:
data == NULL, parameters == NULL). The line above generated a new model, est, which is identical to the base OLS model but has estimated parameters (and covariances, and basic hypothesis tests, a log likelihood,  ,
,  , et cetera), and a
, et cetera), and a data pointer to the apop_data set used for estimation. est. After all, it doesn't make sense to draw from a Normal distribution until its mean and standard deviation are specified. Settings
How many bins are in a histogram? At what tolerance does the maximum likelihood search end? What are the models being combined in an apop_mixture distribution?
Apophenia organizes settings in settings groups, which are then attached to models. In the following snippet demonstrating Bayesian updating, we specify a Beta distribution prior. If the likelihood function were a Binomial distribution, apop_update knows the closed-form posterior for a Beta-Binomial pair, but in this case, with a PMF as a likelihood, it will have to run Markov chain Monte Carlo. The apop_mcmc_settings group attached to the prior specifies details of how the run should work.
For a likelihood, we generate an empirical distribution—a PMF—from an input data set, via apop_estimate(your_data, apop_pmf). When we call apop_update on the last line, it already has all of the above info on hand.
Databases and models
Returning to the introductory example, you saw that (1) the library expects you to keep your data in a database, pulling out the data as needed, and (2) that the workflow is built around apop_model structures.
Starting with (2), if a stats package has something called a model, then it is probably of the form Y = [an additive function of X], such as 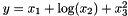 . Trying new models means trying different functional forms for the right-hand side, such as including
. Trying new models means trying different functional forms for the right-hand side, such as including  in some cases and excluding it in others. Conversely, Apophenia is designed to facilitate trying new models in the broader sense of switching out a linear model for a hierarchical, or a Bayesian model for a simulation. A formula syntax makes little sense over such a broad range of models.
in some cases and excluding it in others. Conversely, Apophenia is designed to facilitate trying new models in the broader sense of switching out a linear model for a hierarchical, or a Bayesian model for a simulation. A formula syntax makes little sense over such a broad range of models.
As a result, the right-hand side is not part of the apop_model. Instead, the data is assumed to be correctly formatted, scaled, or logged before being passed to the model. This is where part (1), the database, comes in, because it provides a proxy for the sort of formula specification language above:
Generating factors and dummies is also considered data prep, not model internals. See apop_data_to_dummies and apop_data_to_factors.
Now that you have est, an estimated model, you can interrogate it. This is where Apophenia and its encapsulated model objects shine, because you can do more than just admire the parameter estimates on the screen: you can take your estimated data set and fill in or generate new data, use it as an input to the parent distribution of a hierarchical model, et cetera. Some simple examples:
This introduction has shown you the apop_data set and some of the functions associated, which might be useful even if you aren't formally doing statistical work but do have to deal with data with real-world elements like column names and mixed numeric/text values. You've seen how Apophenia encapsulates many of a model's characteristics into a single apop_model object, which you can send with data to functions like apop_estimate, apop_predict, or apop_draw. Once you've got your data in the right form, you can use this to simply estimate model parameters, or as an input to later analysis.
What's next?
 -,
-,  -distributed statistics); and for the parameters of any model; in Tests & diagnostics.
-distributed statistics); and for the parameters of any model; in Tests & diagnostics.