Here is the model for all hypothesis testing within Apophenia:
There are a handful of named tests that produce a known statistic and then compare to a known distribution, like apop_test_kolmogorov or apop_test_fisher_exact. For traditional distributions (Normal, 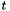 ,
,  ), use the apop_test convenience function.
), use the apop_test convenience function.
In especially common cases, like the parameters from an OLS regression, the commonly-associated 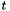 test is included as part of the estimation output, typically as a row in the
test is included as part of the estimation output, typically as a row in the info element of the output apop_model.
See also these Monte Carlo methods:
To give another example of testing, here is a function that was briefly a part of Apophenia, but seemed a bit out of place. Here it is as a sample:
Or, consider the Rao statistic,  where
where  is a model's likelihood function and
is a model's likelihood function and  its information matrix. In code:
its information matrix. In code:
Given the correct assumptions, this is  , where
, where 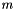 is the dimension of
is the dimension of  , so the odds of a Type I error given the model is:
, so the odds of a Type I error given the model is:
Generalized parameter tests
But if your model is not from the textbook, then you have the tools to apply the above three-step process to the parameters of any apop_model.
apop_estimate(your_data, your_model) will output a model with a parameters element. Defaults for the parameter models are filled in via bootstrapping or resampling, meaning that if your model's parameters are decidedly off the Normal path, you can still test claims about the parameters.
The introductory example in A quick overview ran a standard OLS regression, whose output includes some standard hypothesis tests; to conclude, let us go the long way and replicate those results via the general apop_parameter_model mechanism. The results here will of course be identical, but the more general mechanism can be used in situations where the standard models don't apply.
The first part of this program is identical to the introductory program, using ss08pdc.csv if you have downloaded it as per the instructions in A quick overview, or a simple sample data set if not. The second half executes the three steps uses many of the above features: one of the inputs to apop_parameter_model (which row of the parameter set to use) is sent by adding a settings group, we pull that row into a separate data set using Apop_r, and we set its vector value by referring to it as the -1st element.
Note that the procedure did not assume the model parameters had a certain form. It queried the model for the distribution of parameter agep, and if the model didn't have a closed-form answer then a distribution via bootstrap would be provided. Then that model was queried for its CDF. [The procedure does assume a symmetric distribution. Fixing this is left as an exercise for the reader.] For a model like OLS, this is entirely overkill, which is why OLS provides the basic hypothesis tests automatically. But for models where the distribution of parameters is unknown or has no closed-form solution, this may be the only recourse.