This section is a detailed description of the stock models that ship with Apophenia. It is a reference. For an explanation of what to do with an apop_model, see Models.
The primary questions one has about a model in practice are what format the input data should take and what to expect of an estimated output.
Generally, the input data consists of an apop_data set where each row is a single observation. Details beyond that are listed below.
The output after running apop_estimate to produce a fitted model are generally found in three places: the vector of the output parameter set, its matrix, or a new settings group. The basic intuition is that if the parameters are always a short list of scalars, they are in the vector; if there exists a situation where they could take matrix form, the parameters will be in the matrix; if they require more structure than that, they will be a settings group.
If the basic structure of the apop_data set is unfamiliar to you, see Data sets, which will discuss the basic means of getting data out of a struct. For example, the estimated apop_normal distribution has the mean in position zero of the vector and the standard deviation in position one, so they could be extracted as follows:
See Models for discussion of how to pull settings groups using Apop_settings_get (for one item) or apop_settings_get_group (for a full settings group).
| apop_model apop_bernoulli |
The Bernoulli model: A single random draw with probability 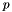 .
.
| apop_model apop_beta |
The beta distribution has two parameters and is restricted to data between zero and one. You may also find apop_beta_from_mean_var to be useful.
| apop_model apop_binomial |
The multi-draw generalization of the Bernoulli, or the two-bin special case of the Multinomial distribution.
It is implemented as an alias of the apop_multinomial model, except that it has an explicit CDF, we know it has two parameters, and its draw method returns a scalar. I.e., .vsize==2 and .dsize==1.
| Input format |
Each row of the matrix is one observation, consisting of two elements. The number of draws of type zero (sometimes read as `misses' or `failures') are in column zero, the number of draws of type one (`hits', `successes') in column one. |
| Parameter format |
a vector, v[0]= |
| Post-estimate data |
Unchanged. |
| RNG |
The RNG returns a single number representing the success count, not a vector of length two giving both the failure bin and success bin. This is notable because it differs from the input data format, but it tends to be what people expect from a Binomial RNG. For draws with both dimensions (or situations where draws are fed back into the model), use an apop_multinomial model with |
| apop_model apop_coordinate_transform |
Apply a coordinate transformation of the data to produce a distribution over the transformed data space. This is sometimes called a Jacobian transformation.
Here is an example that replicates the Lognormal distribution. Note the use of apop_model_copy_set to set up a model with the given settings.
| Name |
|
| Input format |
The input data is sent to the first model, so use the input format for that model. |
| Post-estimate data |
Unchanged. |
| Settings |
| apop_model apop_cross |
A cross product of models. Generate via apop_model_cross .
For the case when you need to bundle two uncorrelated models into one larger model. For example, the prior for a multivariate normal (whose parameters are a vector of means and a covariance matrix) is a Multivariate Normal-Wishart pair.
| apop_model apop_dconstrain |
A model that constrains the base model to within some data constraint. E.g., truncate 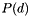 to zero for all
to zero for all 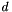 outside of a given constraint. Generate using apop_model_dconstrain .
outside of a given constraint. Generate using apop_model_dconstrain .
The log likelihood works by using the base_model log likelihood, and then scaling it based on the part of the base model's density that is within the constraint. If you have an easy means of specifying what that density is, please do, as in the example. If you do not, the log likelihood will calculate it by making draw_ct random draws from the base model and checking whether they are in or out of the constraint. Because this default method is stochastic, there is some loss of precision.
The previous scaling is stored in the apop_dconstrain settings group. Get/set via:
If scale is zero, because that is the default or because you set it as above, then I recalculate the scale. If the value of the parameters changed since scale was last calculated, I recalculate. If you made other relevant changes to the scale, then you may need to manually zero out scale so it can be recalculated.
Here is an example that makes a few draws and estimations from data-constrained models. Note the use of apop_model_set_settings to prepare the constrained models.
| Name |
|
| Input format |
That of the base model. |
| Parameter format |
That of the base model. In fact, the |
| Post-estimate data |
Unchanged. |
| RNG |
Draw from the base model; if the draw is outside the constraint, throw it out and try again. |
| Settings | |
| Examples |
#include <apop.h> //The constraint function. return apop_data_get(in) > 0; } //The optional scaling function. double in_bounds(apop_model *m){ double z = 0; gsl_vector_view vv = gsl_vector_view_array(&z, 1); } int main(){ /*Set up a Normal distribution, with data truncated to be nonnegative. This version doesn't use the in_bounds function above, and so the default scaling function is used.*/ gsl_rng *r = apop_rng_alloc(213); apop_model *norm = apop_model_set_parameters(apop_normal, 1.2, 0.8); apop_model *trunc = apop_model_set_settings(apop_dconstrain, .base_model=apop_model_copy(norm), .constraint=over_zero, .draw_ct=5e4, .rng=r); //make draws. Currently, you need to prep the model first. apop_prep(NULL, trunc); apop_data *d = apop_model_draws(trunc, 1e5); //Estimate the parameters given the just-produced data: apop_model *est = apop_estimate(d, trunc); apop_model_print(est); assert(apop_vector_distance(est->parameters->vector, norm->parameters->vector)<1e-1); //Generate a data set that is truncated at zero using alternate means apop_data *normald = apop_model_draws(apop_model_set_parameters(apop_normal, 0, 1), 5e4); for (int i=0; i< normald->matrix->size1; i++){ double *d = apop_data_ptr(normald, i); if (*d < 0) *d *= -1; } //this time, use an unparameterized model, and the in_bounds fn apop_model *re_trunc = apop_model_set_settings(apop_dconstrain, .base_model=apop_normal, .constraint=over_zero, .scaling=in_bounds); apop_model *re_est = apop_estimate(normald, re_trunc); apop_model_print(re_est); assert(apop_vector_distance(re_est->parameters->vector, apop_vector_fill(gsl_vector_alloc(2), 0, 1))<1e-1); apop_model_free(trunc); } |
| apop_model apop_dirichlet |
A multivariate generalization of the Beta distribution.
| apop_model apop_exponential |
The Exponential distribution.
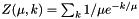

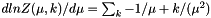
Some write the function as:  If you prefer this form, just convert your parameter via
If you prefer this form, just convert your parameter via 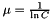 (and convert back from the parameters this function gives you via
(and convert back from the parameters this function gives you via 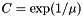 ).
).
| Name |
|
| Input format |
One scalar observation per row (in the |
| Parameter format |
|
| Post-estimate data |
Unchanged. |
| RNG |
Just a wrapper for |
| CDF |
Returns a scalar draw. |
| apop_model apop_gamma |


 (also,
(also,  )
)
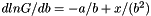
| Name |
|
| Input format |
A scalar, in the See also apop_data_rank_compress for means of dealing with one more input data format. |
| Parameter format |
First two elements of the vector are $$ and $$. |
| Post-estimate data |
Unchanged. |
| RNG |
A wrapper for See the notes for apop_exponential on a popular alternate form. |
| apop_model apop_improper_uniform |
The improper uniform returns 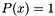 for every value of x, all the time (and thus, log likelihood(x)=0). It has zero parameters.
for every value of x, all the time (and thus, log likelihood(x)=0). It has zero parameters.
| apop_model apop_iv |
Instrumental variable regression
Operates much like the apop_ols model, but the input parameters also need to have a table of substitutions (like the addition of the .instruments setting in the example below).
Which columns substitute where can be specified in your choice of two ways. The first is to use the vector element of the apop_data set to list the column numbers to be substituted (the dependent variable is zero; first independent column is one), and then one column for each item to substitute.
The second method, if the vector of the instrument apop_data set is NULL, is to use the column names to find the matching columns in the base data to substitute. This is generally more robust and/or convenient.
instruments data set is NULL or empty, I'll just run OLS.destroy_data setting. If you set that to 'y', I will overwrite the column in place, saving the trouble of copying the entire data set. | Name |
|
| Input format |
See the discussion on the apop_ols page regarding its prep routine. See above regarding the |
| Prep routine |
See the discussion on the apop_ols page regarding its prep routine. |
| Parameter format |
As per apop_ols |
| Post-estimate data |
Unchanged. |
| Examples |
/* Instrumental variables are often used to deal with variables measured with noise, so this example produces a data set with a column of noisy data, and a separate instrument measured with greater precision, then sets up and runs an instrumental variable regression. To guarantee that the base data set has noise and the instrument is cleaner, the procedure first generates the clean data set, then copies the first column to the instrument set, then the add_noise function inserts Gaussian noise into the base data set. Once the base set and the instrument set have been generated, the setup for the IV consists of adding the relevant names and using Apop_model_add_group to add a lm (linear model) settings group with an .instrument=instrument_data element. In fact, the example sets up a sequence of IV regressions, with more noise each time. */ #include <apop.h> #define Diff(L, R, eps) Apop_stopif(fabs((L)-(R)>=(eps)), return, 0, "%g is too different \ from %g (abitrary limit=%g).", (double)(L), (double)(R), eps); int datalen =1e4; //generate a vector that is the original vector + noise void add_noise(gsl_vector *in, gsl_rng *r, double size){ apop_model *nnoise = apop_model_set_parameters(apop_normal, 0, size); apop_data *nd = apop_model_draws(nnoise, in->size); gsl_vector_add(in, Apop_cv(nd, 0)); /*for (int i=0; i< in->size; i++){ double noise; apop_draw(&noise, r, nnoise); *gsl_vector_ptr(in, i) += noise; }*/ apop_data_free(nd); apop_model_free(nnoise); } Diff(apop_data_get(m->parameters, 0, -1), -1.4, tolerance); Diff(apop_data_get(m->parameters, 1, -1), 2.3, tolerance); } int main(){ gsl_rng *r = apop_rng_alloc(234); apop_data *data = apop_data_alloc(datalen, 2); for(int i=0; i< datalen; i++){ apop_data_set(data, i, 1, 100*(gsl_rng_uniform(r)-0.5)); apop_data_set(data, i, 0, -1.4 + apop_data_get(data,i,1)*2.3); } apop_model_show(oest); //the data with no noise will be the instrument. gsl_vector *col1 = Apop_cv(data, 1); apop_data *instrument_data = apop_data_alloc(data->matrix->size1, 1); gsl_vector_memcpy(Apop_cv(instrument_data, 0), col1); Apop_model_add_group(apop_iv, apop_lm, .instruments = instrument_data); //Now add noise to the base data four times, and estimate four IVs. int tries = 4; apop_model *ests[tries]; for (int nscale=0; nscale<tries; nscale++){ add_noise(col1, r, nscale==0 ? 0 : pow(10, nscale-tries)); ests[nscale] = apop_estimate(data, apop_iv); if (nscale==tries-1){ //print the one with the largest error. printf("\nnow IV:\n"); apop_model_show(ests[nscale]); } } /* Now test. The parameter estimates are unbiased. As we add more noise, the covariances expand. Test that the ratio of one covariance matrix to the next is less than one, though these are typically very much smaller than one (as the noise is an order of magnitude larger in each case), and the ratios will be identical for each j, k below. */ test_for_unbiased_parameter_estimates(ests[0], 1e-6); for (int i=1; i<tries; i++){ test_for_unbiased_parameter_estimates(ests[i], 1e-3); gsl_matrix *cov = apop_data_get_page(ests[i-1]->parameters, "<Covariance>")->matrix; gsl_matrix *cov2 = apop_data_get_page(ests[i]->parameters, "<Covariance>")->matrix; gsl_matrix_div_elements(cov, cov2); for (int j =0; j< 2; j++) for (int k =0; k< 2; k++) assert(gsl_matrix_get(cov, j, k) < 1); } } |
| apop_model apop_kernel_density |
The kernel density smoothing of a PMF or histogram.
At each point along the histogram, put a distribution (default: Normal(0,1)) on top of the point. Sum all of these distributions to form the output distribution.
Setting up a kernel density consists of setting up a model with the base data and the information about the kernel model around each point. This can be done using the apop_model_set_settings function to get a copy of the base apop_kernel_density model and add a apop_kernel_density_settings group with the appropriate information; see the main function of the example below.
| Name |
|
| Input format |
One observation per line. Each row in turn will be passed through to the elements of |
| Parameter format |
None |
| Post-estimate data |
Unchanged. |
| RNG |
Randomly selects a data point, then randomly draws from that sub-distribution. Returns 0 on success, 1 if unable to pick a sub-distribution (meaning the weights over the distributions are somehow broken), and 2 if unable to draw from the sub-distribution. |
| CDF |
Sums the CDF to the given point of all the sub-distributions. |
| Settings |
apop_kernel_density_settings, including:
See the sample code for for a Uniform[0,1] recentered around the first element of the PMF matrix. |
| Examples |
This example sets up and uses KDEs based on Normal and Uniform distributions. /* This program draws ten random data points, and then produces two kernel density estimates: one based on the Normal distribution and one based on the Uniform. It produces three outputs: --stderr shows the random draws --kerneldata is a file written with plot data for both KDEs --stdout shows instructions to gnuplot, so you can pipe: ./kernel | gnuplot -persist Most of the code is taken up by the plot() and draw_some_data() functions, which are straightforward. Notice how plot() pulls the values of the probability distributions at each point along the scale. The set_uniform_edges function sets the max and min of a Uniform distribution so that the given point is at the center of the distribution. The first KDE uses the defaults, which are based on a Normal distribution with std dev 1; the second explicitly sets the .kernel and .set_fn for a Uniform. */ #include <apop.h> apop_data_set(unif->parameters, 0, -1, r->matrix->data[0]-0.5); apop_data_set(unif->parameters, 1, -1, r->matrix->data[0]+0.5); } apop_data *onept = apop_data_alloc(1,1); FILE *outtab = fopen("kerneldata", "w"); for (double i=0; i<20; i+=0.01){ apop_data_set(onept, .val=i); } fclose(outtab); printf("plot 'kerneldata' using 1:2\n" "replot 'kerneldata' using 1:3\n"); } apop_data *draw_some_data(){ apop_model *uniform_0_20 = apop_model_set_parameters(apop_uniform, 0, 20); apop_data *d = apop_model_draws(uniform_0_20, 10); apop_data_print(apop_data_sort(d), .output_pipe=stderr); return d; } int main(){ apop_data *d = draw_some_data(); apop_model *k2 = apop_model_set_settings(apop_kernel_density, .base_data=d, .set_fn = set_uniform_edges, .kernel = apop_uniform); plot(k, k2); } |
| apop_model apop_loess |
Regression via loess smoothing
This uses a somewhat black-box routine, first written by Chamberlain, Devlin, Grosse, and Shyu in 1988, to fit a smoothed series of quadratic curves to the input data, thus producing a curve more closely fitting than a simple regression would.
The curve is basically impossible to describe using a short list of parameters, so the representation is in the form of the predicted vector of the expected data set; see below.
From the 1992 manual for the package: ``The method we will use to fit local regression models is called loess, which is short for local regression, and was chosen as the name since a loess is a deposit of fine clay or silt along a river valley, and thus is a surface of sorts. The word comes from the German löss, and is pronounced löíss.''
| Name |
|
| Input format |
The data is basically OLS-like: the first column of the data is the dependent variable to be explained; subsequent variables are the independent explanatory variables. Thus, your input data can either have a dependent vector plus explanatory matrix, or a matrix where the first column is the dependent variable. Unlike with OLS, I won't move your original data, and I won't add a 1, because that's not really the loess custom. You can of course set up your data that way if you like. If your data set has a weights vector, I'll use it. In any case, all data is copied into the model's apop_loess_settings. The code is primarily FORTRAN code from 1988 converted to C; the data thus has to be converted into a relatively obsolete internal format. |
| Parameter format |
Unused. |
| Post-estimate data |
Unchanged. |
| Post-estimate parameters |
None. |
| Predict |
Fills in the zeroth column (ignoring and overwriting any data there), and adds an additional page to the input apop_data set named "<Confidence>" with a lower and upper CI for each point. |
| apop_model apop_logit |
Apophenia makes no distinction between the bivariate logit and the multinomial logit. This does both.
The likelihood of choosing item  is:
is: 
so the log likelihood is 
| Name |
|
| Input format |
The first column of the data matrix this model expects is zeros, ones, ..., enumerating the factors; to get there, try apop_data_to_factors; if you forget to run it, I'll run it on the first data column for you. The remaining columns are values of the independent variables. Thus, the model will return [(data columns)-1] |
| Prep routine |
You will probably want to convert some column of your data into factors, via apop_data_to_factors. If you do, then that adds a page of factors to your data set (and of course adjusts the data itself). If I find a factor page, I will use that info; if not, then I will run apop_data_to_factors on the first column (the vector if there is one, else the first column of the matrix.) Also, if there is no vector, then I will move the first column of the matrix, and replace that matrix column with a constant column of ones, just like with OLS. |
| Parameter format |
As above. |
| Post-estimate data |
Unchanged. |
| RNG |
Much like the apop_ols RNG, qv. Returns the category drawn. Here is an artifical example which clarifies the simplest use of the model: #include <apop.h> #include <unistd.h> char *testfile = "logit_test_data"; //generate a fake data set. //Notice how the first column is the outcome, just as with standard regression. void write_data(){ FILE *f = fopen(testfile, "w"); fprintf(f, "\ outcome,A, B \n\ 0, 0, 0 \n\ 1, 1, 1 \n\ 1, .7, .5 \n\ 1, .7, .3 \n\ 1, .3, .7 \n\ \n\ 1, .5, .5 \n\ 0, .4, .4 \n\ 0, .3, .4 \n\ 1, .1, .3 \n\ 1, .3, .1 "); fclose(f); } int main(){ write_data(); apop_data *d = apop_text_to_data(testfile); Apop_model_add_group(apop_logit, apop_mle, .tolerance=1e-5); unlink(testfile); /* Apophenia's test suite checks that this code produces values close to canned values. As a human, you probably just want to print the results to the screen. */ apop_model_show(est); assert(fabs(apop_data_get(est->parameters, .rowname="1")- -1.155026) < 1e-6); assert(fabs(apop_data_get(est->parameters, .rowname="A")- 4.039903) < 1e-6); assert(fabs(apop_data_get(est->parameters, .rowname="B")- 1.494694) < 1e-6); } Here is an example using data from a U.S. Congressional vote, including one text variable that has to be converted to factors, and one to convert to dummies. A loop then calculates the customary p-values. // See http://modelingwithdata.org/arch/00000160.htm for context and analysis. #ifdef Datadir #define DATADIR Datadir #else #define DATADIR "." #endif #include <apop.h> int main(){ //read the data to db, get the desired columns, //prep the two categorical variables apop_data *d = apop_query_to_mixed_data("mmmtt", "select 0, ideology,log(contribs+10) as contribs, vote, party from amash"); apop_data_to_factors(d); //0th text col -> 0th matrix col //Estimate a logit model, get covariances, //calculate p values under popular Normality assumptions Apop_model_add_group(apop_logit, apop_parts_wanted, .covariance='y'); apop_model_show(out); for (int i=0; i< out->parameters->matrix->size1; i++){ printf("%s pval:\t%g\n",out->parameters->names->row[i], apop_test(apop_data_get(out->parameters, i), "normal", 0, sqrt(apop_data_get(out->parameters->more, i, i)))); } } |
| apop_model apop_lognormal |
The log likelihood function for lognormal distributions:


| Name |
|
| Input format |
A scalar in the the matrix or vector element of the input apop_data set. |
| Parameter format |
Zeroth vector element is the mean of the logged data set; first is the standard deviation of the logged data set. |
| Post-estimate data |
Unchanged. |
| Post-estimate info |
Reports |
| RNG |
An Apophenia wrapper for the GSL's Normal RNG, exponentiated. |
| apop_model apop_mixture |
The mixture model transformation: a linear combination of multiple models.
Use apop_model_mixture to produce one of these models. In the examples below, some are generated from unparameterized input models with a form like
Or, one can skip the estimation and use already-parameterized models as input to apop_model_mixture, e.g.:
Notice that the weights vector has to be added after the call to apop_model_mixture. If none is given, then equal weights are assigned to all components of the mixture.
One can think of the estimation in the un-parameterized case as a missing-data problem: each data point originated in one distribution or the other, and if we knew with certainty which data point came from which distribution, then the estimation problem would be trivial: just generate the subsets and call apop_estimate(dataset1, model1), ..., apop_estimate(datasetn, modeln) separately. But the assignment of which element goes where is unknown information, which we guess at using an expectation-maximization (EM) algorithm. The standard algorithm starts with an initial set of parameters for the models, and assigns each data point to its most likely model. It then re-estimates the model parameters using their subsets. The standard algorithm, see e.g. this PDF, repeats until it arrives at an optimum.
Thus, the log likelihood method for this model includes a step that allocates each data point to its most likely model, and calculates the log likelihood of each observation using its most likely model. [It would be a valuable extension to extend this to not-conditionally IID models. Commit 1ac0dd44 in the repository had some notes on this, now removed.] As a side-effect, it calculates the odds of drawing from each model (the vector λ). Following the above-linked paper, the probability for a given observation under the mixture model is its probability under the most likely model weighted by the previously calculated  for the given model.
for the given model.
Apophenia implements the EM algorithm as a constrained optimization(!). The constraint check repositions the vector of weights to that calculated at the last step, then the log likelihood calculates the likelihood as above, including the expected value of the weights vector for the next step. Thus, Apophenia casts the Expectation step as a step repositioning the maximization's constraint and its associated penalties.
Estimations of mixture distributions can be sensitive to initial conditions. You are encouraged to try a sequence of random starting points for your model parameters. Some authors recommend plotting the data and eyeballing a guess as to the model parameters.
| Name |
|
| Input format |
The same data gets sent to each of the component models of the mixture. Each row is an observation, and the estimation routine assumes that models are conditionally IID (i.e., having chosen what component of the mixture the observation comes from, its likelihood can be calculated independently of all other observations). |
| Parameter format |
The parameters are broken out in a readable form in the settings group, so your best bet is to use those. See the sample code for usage. |
| Post-estimate data |
Unchanged. |
| RNG |
Uses the weights to select a component model, then makes a draw from that component. The model's |
| Settings | |
| Examples |
The first example uses a text file #ifdef Datadir #define DATADIR Datadir #else #define DATADIR "." #endif #include <apop.h> /* This replacement for apop_model_print(in) demonstrates retrieval of the useful settings: the weights (λ) and list of estimated models. It is here only for demonstration purposes---it is what apop_model_print(your_mix) will do. */ void show_mix(apop_model *in){ printf("The weights:\n"); apop_vector_print(ms->weights); printf("\nThe models:\n"); apop_model_print(*m, stdout); } int main(){ apop_data *dd = apop_query_to_data("select waiting from ff"); Apop_settings_set(mf, apop_mixture, find_weights, 'y');//Use the EM algorithm to search for optimal weights. /* The process is famously sensitive to starting points. Try many random points, or eyeball the distribution's plot and guess at the starting values. */ Apop_model_add_group(mf, apop_mle, .starting_pt=(double[]){.5, .5, 50, 5, 80, 5}, .step_size=3, .tolerance=1e-6); apop_model *mfe = apop_estimate(dd, mf); apop_model_print(mfe, stdout); printf("LL=%g\n", apop_log_likelihood(dd, mfe)); printf("\n\nValues calculated in the source paper, for comparison.\n"); apop_model *r_ed = apop_model_mixture( apop_model_set_parameters(apop_normal, 54.61364, 5.869089), apop_model_set_parameters(apop_normal, 80.09031, 5.869089)); apop_data *wts = apop_data_falloc((2), 0.3608498, 0.6391502); Apop_settings_add(r_ed, apop_mixture, weights, wts->vector); show_mix(r_ed); printf("LL=%g\n", apop_log_likelihood(dd, r_ed)); } This example begins with a fixed mixture distribution, and makes assertions about the characteristics of draws from it. #include <apop.h> /* Use apop_model_mixture to generate a hump-filled distribution, then find the most likely data points and check that they are near the humps. */ //Produce a 2-D multivariate normal model with unit covariance and given mean out->parameters = apop_data_falloc((2, 2, 2), x, 1, 0, y, 0, 1); out->dsize = 2; return out; } int main(){ //here's a mean/covariance matrix for a standard multivariate normal. apop_model *many_humps = apop_model_mixture( produce_fixed_mvn(5, 6), produce_fixed_mvn(-5, -4), produce_fixed_mvn(0, 1)); apop_prep(NULL, many_humps); int len = 100000; apop_data *d = apop_model_draws(many_humps, len); gsl_vector *first = Apop_cv(d, 0); printf("mu=%g\n", apop_mean(first)); assert(fabs(apop_mean(first)- 0) < 5e-2); gsl_vector *second = Apop_cv(d, 1); printf("mu=%g\n", apop_mean(second)); assert(fabs(apop_mean(second)- 1) < 5e-2); /* Use the ML imputation routine to search for the input value with the highest log likelihood. Do the search via simulated annealing. */ apop_data *x = apop_data_alloc(1,2); gsl_matrix_set_all(x->matrix, NAN); apop_opts.stop_on_warning='v'; apop_ml_impute(x, many_humps); printf("Optimum found at:\n"); apop_data_show(x); assert(fabs(apop_data_get(x, .col=0)- 0) + fabs(apop_data_get(x, .col=1) - 1) < 1e-2); } |
| apop_model apop_multinomial |
The 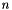 –option generalization of the Binomial distribution.
–option generalization of the Binomial distribution.
| Name |
|
| Input format |
Each row of the matrix is one observation: a set of draws from a single bin. The number of draws of type zero are in column zero, the number of draws of type one in column one, et cetera.
|
| Parameter format |
The parameters are kept in the vector element of the The numeraire is bin zero, meaning that And now the parameter vector is a proper list of probabilities.
|
| Post-estimate data |
Unchanged. |
| Post-estimate info |
Reports |
| RNG |
Returns a single vector of length |
| apop_model apop_multivariate_normal |
This is the multivariate generalization of the Normal distribution.
| Name |
|
| Input format |
Each row of the matrix is an observation. |
| Parameter format |
An If you had only one dimension, the mean would be a vector of size one, and the covariance matrix a After estimation, the |
| Post-estimate data |
Unchanged. |
| Post-estimate info |
Reports |
| RNG |
From Devroye (1986), p 565. |
| apop_model apop_normal |
You know it, it's your attractor in the limit, it's the Gaussian distribution.


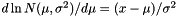
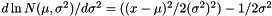
See also the apop_multivariate_normal.
| Name |
|
| Input format |
A scalar, in the |
| Parameter format |
Parameter zero (in the vector) is the mean, parmeter one is the standard deviation (i.e., the square root of the variance). After estimation, a page is added named |
| Post-estimate data |
Unchanged. |
| Post-estimate info |
Reports the log likelihood. |
| Predict |
|
| RNG |
A wrapper for the GSL's Normal RNG. |
| Settings |
None. |
| apop_model apop_ols |
Ordinary least squares. Weighted least squares is also handled by this model.
| Name |
|
| Input format |
See the notes on the prep routine. If you provide weights in |
| Prep routine |
If your input data has no If your data has a |
| Parameter format |
A vector of OLS coefficients. Coefficient zero refers to the constant column, if any. The The estimation routine appends a page to the |
| Post-estimate data |
You can specify whether the data is modified with an apop_lm_settings group. Else, left unchanged. |
| Post-estimate parameter model |
For the mean, a noncentral |
| Post-estimate info |
Reports log likelihood, and runs apop_estimate_coefficient_of_determination to add Residuals: I add a page named Given your estimate |
| RNG |
Linear models are typically only partially defined probability models. For OLS, we know that The apop_lm_settings group includes an apop_model element named The default is that But you can't draw from an improper uniform. So if you draw from a linear model with a default Alternatively, you may know something about the distribution of the input data. For example, the data model may simply be a PMF from the actual data: Now, random draws are taken from the input data, and the dependent variable value calculated via |
| Examples |
A quick overview opens with a sample program using OLS. For quick reference, here is the program, but see that page for a full discussion. #ifdef Datadir #define DATADIR Datadir #else #define DATADIR "." #endif #include <apop.h> int main(){ apop_data *data = apop_query_to_data("select * from d"); apop_model_print(est); } |
| apop_model apop_pmf |
A probability mass function is commonly known as a histogram, or still more commonly, a bar chart. It indicates that at a given coordinate, there is a given mass.
Each row of the PMF's data set holds the coordinates, and the weights vector holds the mass at the given point. This is in contrast to the crosstab format, where the location is simply given by the position of the data point in the grid.
For example, here is a typical crosstab:
| col 0 | col 1 | col 2 | |
| row 0 | 0 | 8.1 | 3.2 |
| row 1 | 0 | 0 | 2.2 |
| row 2 | 0 | 7.3 | 1.2 |
Here it is as a sparse listing:
| dimension 1 | dimension 2 | value |
| 0 | 1 | 8.1 |
| 0 | 2 | 3.2 |
| 1 | 2 | 2.2 |
| 2 | 1 | 7.3 |
| 2 | 2 | 1.2 |
The apop_pmf internally represents data in this manner, with the dimensions in the matrix, vector, and text element of the data set, and the cell values are held in the weights element (not the vector).
If your data is in a crosstab (with observation coordinates in the matrix element for 2-D data or the vector for 1-D data), then use apop_crosstab_to_db to make the conversion. See also the wiki for another crosstab-to-PMF function.
If your data is already in the sparse listing format (which is probably the case for 3- or more dimensional data), then estimate the model via:
weights element is NULL, then I assume that all rows of the data set are equally probable. weights are present but sum to a not-finite value, the model's error element is set to 'w' when the estimation is run, and a warning printed. | Name |
| ||||
| Input format |
One observation per row, with coordinates in the | ||||
| Parameter format |
None. The list of observations and their weights are in the | ||||
| Post-estimate data |
The data you sent in is linked to (not copied). | ||||
| Post-estimate parameters |
Still | ||||
| RNG |
Return the data in a random row of the PMF's data set. If there is a weights vector, I will use that to make draws; else all rows are equiprobable.
then I will return the row number of the draw, not the data in that row. Because apop_draw only returns numeric data, this is the only meaningful way to make draws from text data.
| ||||
| CDF |
Assuming the data is sorted in a meaningful manner, find the total mass up to a given data point. That is, a CDF only makes sense if the data space is totally ordered. The sorting you define using apop_data_sort defines that ordering.
| ||||
| Settings |
| apop_model apop_poisson |
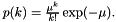
| Name |
|
| Input format |
One scalar observation per row (in the |
| Parameter format |
One parameter, the zeroth element of the vector ( |
| Post-estimate data |
Unchanged. |
| Post-estimate parameters |
Unless you decline it by adding the apop_parts_wanted_settings group, I will also give you the variance of the parameter, via bootstrap, stored in a page named |
| Post-estimate info |
Reports |
| RNG |
A wrapper for |
| apop_model apop_probit |
Apophenia makes no distinction between the Bivariate Probit and the Multinomial Probit. This one does both.
| Name |
|
| Input format |
The first column of the data matrix this model expects is zeros, ones, ..., enumerating the factors; see the prep routine. The remaining columns are values of the independent variables. Thus, the model will return [(data columns)-1] |
| Prep routine |
The initial column of data should be a set of factors, set up via apop_data_to_factors. If I find a factor page, I will use that info; if not, then I will run apop_data_to_factors on the left-most column (the vector if there is one, else the first column of the matrix.) Also, if there is no vector, then I will move the first column of the matrix, and replace that matrix column with a constant column of ones, just like with OLS. |
| Parameter format |
As above |
| Post-estimate data |
Unchanged. |
| RNG |
See apop_ols; this one is similar but produces a category number instead of OLS's continuous draw. |
| apop_model apop_t_distribution |
The t distribution, primarily for descriptive purposes.
If you want to test a hypothesis, you probably don't need this, and should instead use apop_test.
In that world, the 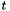 distribution is parameter free. The data are assumed to be normalized to be based on a mean zero, variance one process, you get the degrees of freedom from the size of the data, and the distribution is thus fixed.
distribution is parameter free. The data are assumed to be normalized to be based on a mean zero, variance one process, you get the degrees of freedom from the size of the data, and the distribution is thus fixed.
For modeling purposes, more could be done. For example, the t-distribution is a favorite proxy for Normal-like situations where there are fat tails relative to the Normal (i.e., high kurtosis). Or, you may just prefer not to take the step of normalizing your data—one could easily rewrite the theorems underlying the t-distribution without the normalizations.
In such a case, the researcher would not want to fix the 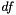 , because
, because 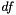 indicates the fatness of the tails, which has some optimal value given the data. Thus, there are two modes of use for these distributions:
indicates the fatness of the tails, which has some optimal value given the data. Thus, there are two modes of use for these distributions:
—will return exactly the type of 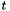 -distribution one would use for testing.
-distribution one would use for testing.
estimate method— 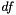 via maximum likelihood, which may be desirable for to find the best-fitting model for descriptive purposes.
via maximum likelihood, which may be desirable for to find the best-fitting model for descriptive purposes. | apop_model apop_uniform |
This is the two-parameter version of the Uniform, expressing a uniform distribution over [a, b].
The MLE of this distribution is simply a = min(your data); b = max(your data). Often useful for the RNG, such as when you have a Uniform prior model.
| apop_model apop_yule |



| Name |
|
| Input format |
One scalar observation per row (in the |
| Parameter format |
One element in the parameter set's vector. |
| Post-estimate data |
Unchanged. |
| RNG |
From Devroye (1986), p 553. |
| Settings |
MLE-type: apop_mle_settings, apop_parts_wanted_settings |
| apop_model apop_zipf |
Wikipedia has notes on the Zipf distribution.

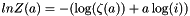
| Name |
|
| Input format |
One scalar observation per row (in the See also apop_data_rank_compress for means of dealing with one more input data format. |
| Parameter format |
One item in the parameter set's vector. |
| Post-estimate data |
Unchanged. |
| RNG |
Returns an ordinal ranking, starting from 1. From Devroye (1986), Chapter 10, p 551. |
| Settings |